| Motif | NKX21.H12INVIVO.0.P.B |
| Gene (human) | NKX2-1 (GeneCards) |
| Gene synonyms (human) | NKX2A, TITF1, TTF1 |
| Gene (mouse) | Nkx2-1 |
| Gene synonyms (mouse) | Nkx-2.1, Titf1, Ttf1 |
| LOGO | 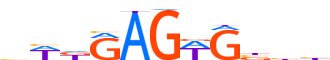 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | NKX21.H12INVIVO.0.P.B |
| Gene (human) | NKX2-1 (GeneCards) |
| Gene synonyms (human) | NKX2A, TITF1, TTF1 |
| Gene (mouse) | Nkx2-1 |
| Gene synonyms (mouse) | Nkx-2.1, Titf1, Ttf1 |
| LOGO | 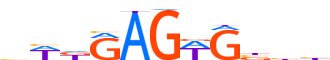 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 11 |
| Consensus | bWdRAGWGbhh |
| GC content | 50.61% |
| Information content (bits; total / per base) | 7.681 / 0.698 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 8 (56) | 0.833 | 0.913 | 0.707 | 0.821 | 0.742 | 0.822 | 1.96 | 2.461 | 115.012 | 293.398 |
| Mouse | 3 (19) | 0.915 | 0.933 | 0.814 | 0.852 | 0.828 | 0.848 | 2.392 | 2.608 | 206.854 | 288.824 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | NK2.1 {3.1.2.14} (TFClass) |
| TFClass ID | TFClass: 3.1.2.14.1 |
| HGNC | HGNC:11825 |
| MGI | MGI:108067 |
| EntrezGene (human) | GeneID:7080 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21869 (SSTAR profile) |
| UniProt ID (human) | NKX21_HUMAN |
| UniProt ID (mouse) | NKX21_MOUSE |
| UniProt AC (human) | P43699 (TFClass) |
| UniProt AC (mouse) | P50220 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 8 human, 3 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | NKX21.H12INVIVO.0.P.B.pcm |
| PWM | NKX21.H12INVIVO.0.P.B.pwm |
| PFM | NKX21.H12INVIVO.0.P.B.pfm |
| Alignment | NKX21.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | NKX21.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | NKX21.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | NKX21.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | NKX21.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 132.0 | 407.0 | 234.0 | 227.0 |
| 02 | 221.0 | 117.0 | 42.0 | 620.0 |
| 03 | 111.0 | 86.0 | 290.0 | 513.0 |
| 04 | 174.0 | 65.0 | 757.0 | 4.0 |
| 05 | 949.0 | 1.0 | 13.0 | 37.0 |
| 06 | 25.0 | 11.0 | 963.0 | 1.0 |
| 07 | 325.0 | 3.0 | 94.0 | 578.0 |
| 08 | 108.0 | 15.0 | 842.0 | 35.0 |
| 09 | 54.0 | 326.0 | 411.0 | 209.0 |
| 10 | 167.0 | 325.0 | 148.0 | 360.0 |
| 11 | 169.0 | 283.0 | 134.0 | 414.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.132 | 0.407 | 0.234 | 0.227 |
| 02 | 0.221 | 0.117 | 0.042 | 0.62 |
| 03 | 0.111 | 0.086 | 0.29 | 0.513 |
| 04 | 0.174 | 0.065 | 0.757 | 0.004 |
| 05 | 0.949 | 0.001 | 0.013 | 0.037 |
| 06 | 0.025 | 0.011 | 0.963 | 0.001 |
| 07 | 0.325 | 0.003 | 0.094 | 0.578 |
| 08 | 0.108 | 0.015 | 0.842 | 0.035 |
| 09 | 0.054 | 0.326 | 0.411 | 0.209 |
| 10 | 0.167 | 0.325 | 0.148 | 0.36 |
| 11 | 0.169 | 0.283 | 0.134 | 0.414 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.633 | 0.485 | -0.066 | -0.096 |
| 02 | -0.122 | -0.752 | -1.75 | 0.904 |
| 03 | -0.803 | -1.054 | 0.147 | 0.715 |
| 04 | -0.359 | -1.328 | 1.103 | -3.783 |
| 05 | 1.329 | -4.525 | -2.839 | -1.872 |
| 06 | -2.243 | -2.985 | 1.344 | -4.525 |
| 07 | 0.261 | -3.975 | -0.967 | 0.834 |
| 08 | -0.83 | -2.711 | 1.209 | -1.925 |
| 09 | -1.508 | 0.264 | 0.494 | -0.178 |
| 10 | -0.4 | 0.261 | -0.52 | 0.363 |
| 11 | -0.388 | 0.123 | -0.618 | 0.502 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.169665 |
| 0.0005 | 5.820935 |
| 0.0001 | 7.03679 |