| Motif | NFIX.H12INVITRO.0.SM.B |
| Gene (human) | NFIX (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Nfix |
| Gene synonyms (mouse) | |
| LOGO | 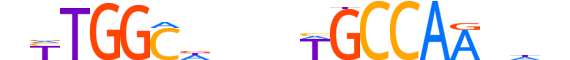 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | NFIX.H12INVITRO.0.SM.B |
| Gene (human) | NFIX (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Nfix |
| Gene synonyms (mouse) | |
| LOGO | 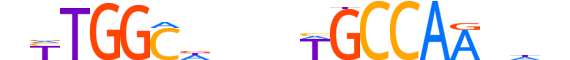 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 19 |
| Consensus | nYTGGCdnndWGCCAAndn |
| GC content | 49.56% |
| Information content (bits; total / per base) | 18.186 / 0.957 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9872 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Methyl HT-SELEX, 1 experiments | median | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 | 0.999 |
| best | 1.0 | 1.0 | 1.0 | 0.999 | 0.999 | 0.999 | |
| Non-Methyl HT-SELEX, 5 experiments | median | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.981 | 0.893 | 0.948 | 0.581 |
| batch 2 | 0.717 | 0.589 | 0.716 | 0.472 |
| TF superclass | beta-Hairpin exposed by an alpha/beta-scaffold {7} (TFClass) |
| TF class | SMAD/NF-1 DNA-binding domain factors {7.1} (TFClass) |
| TF family | NF-1 {7.1.2} (TFClass) |
| TF subfamily | {7.1.2.0} (TFClass) |
| TFClass ID | TFClass: 7.1.2.0.4 |
| HGNC | HGNC:7788 |
| MGI | MGI:97311 |
| EntrezGene (human) | GeneID:4784 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18032 (SSTAR profile) |
| UniProt ID (human) | NFIX_HUMAN |
| UniProt ID (mouse) | NFIX_MOUSE |
| UniProt AC (human) | Q14938 (TFClass) |
| UniProt AC (mouse) | P70257 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 5 |
| Methyl-HT-SELEX | 1 |
| PCM | NFIX.H12INVITRO.0.SM.B.pcm |
| PWM | NFIX.H12INVITRO.0.SM.B.pwm |
| PFM | NFIX.H12INVITRO.0.SM.B.pfm |
| Alignment | NFIX.H12INVITRO.0.SM.B.words.tsv |
| Threshold to P-value map | NFIX.H12INVITRO.0.SM.B.thr |
| Motif in other formats | |
| JASPAR format | NFIX.H12INVITRO.0.SM.B_jaspar_format.txt |
| MEME format | NFIX.H12INVITRO.0.SM.B_meme_format.meme |
| Transfac format | NFIX.H12INVITRO.0.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2183.75 | 1960.75 | 2307.75 | 3419.75 |
| 02 | 857.25 | 1535.25 | 585.25 | 6894.25 |
| 03 | 53.0 | 35.0 | 176.0 | 9608.0 |
| 04 | 38.0 | 22.0 | 9786.0 | 26.0 |
| 05 | 85.0 | 34.0 | 9710.0 | 43.0 |
| 06 | 1485.0 | 8269.0 | 58.0 | 60.0 |
| 07 | 5181.0 | 647.0 | 1815.0 | 2229.0 |
| 08 | 2701.0 | 2987.0 | 1308.0 | 2876.0 |
| 09 | 2720.0 | 1982.0 | 2075.0 | 3095.0 |
| 10 | 2426.0 | 1125.0 | 3400.0 | 2921.0 |
| 11 | 1134.0 | 1054.0 | 135.0 | 7549.0 |
| 12 | 3.0 | 0.0 | 9523.0 | 346.0 |
| 13 | 4.0 | 9840.0 | 20.0 | 8.0 |
| 14 | 2.0 | 9856.0 | 5.0 | 9.0 |
| 15 | 9854.0 | 6.0 | 0.0 | 12.0 |
| 16 | 7925.0 | 21.0 | 1863.0 | 63.0 |
| 17 | 2799.0 | 2445.0 | 2405.0 | 2223.0 |
| 18 | 4131.25 | 940.25 | 1316.25 | 3484.25 |
| 19 | 3331.25 | 1992.25 | 1728.25 | 2820.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.221 | 0.199 | 0.234 | 0.346 |
| 02 | 0.087 | 0.156 | 0.059 | 0.698 |
| 03 | 0.005 | 0.004 | 0.018 | 0.973 |
| 04 | 0.004 | 0.002 | 0.991 | 0.003 |
| 05 | 0.009 | 0.003 | 0.984 | 0.004 |
| 06 | 0.15 | 0.838 | 0.006 | 0.006 |
| 07 | 0.525 | 0.066 | 0.184 | 0.226 |
| 08 | 0.274 | 0.303 | 0.132 | 0.291 |
| 09 | 0.276 | 0.201 | 0.21 | 0.314 |
| 10 | 0.246 | 0.114 | 0.344 | 0.296 |
| 11 | 0.115 | 0.107 | 0.014 | 0.765 |
| 12 | 0.0 | 0.0 | 0.965 | 0.035 |
| 13 | 0.0 | 0.997 | 0.002 | 0.001 |
| 14 | 0.0 | 0.998 | 0.001 | 0.001 |
| 15 | 0.998 | 0.001 | 0.0 | 0.001 |
| 16 | 0.803 | 0.002 | 0.189 | 0.006 |
| 17 | 0.284 | 0.248 | 0.244 | 0.225 |
| 18 | 0.418 | 0.095 | 0.133 | 0.353 |
| 19 | 0.337 | 0.202 | 0.175 | 0.286 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.122 | -0.23 | -0.067 | 0.326 |
| 02 | -1.056 | -0.474 | -1.436 | 1.027 |
| 03 | -3.799 | -4.193 | -2.629 | 1.358 |
| 04 | -4.116 | -4.622 | 1.377 | -4.469 |
| 05 | -3.343 | -4.22 | 1.369 | -3.999 |
| 06 | -0.507 | 1.208 | -3.713 | -3.68 |
| 07 | 0.741 | -1.336 | -0.307 | -0.102 |
| 08 | 0.09 | 0.191 | -0.634 | 0.153 |
| 09 | 0.097 | -0.219 | -0.173 | 0.226 |
| 10 | -0.017 | -0.785 | 0.32 | 0.168 |
| 11 | -0.777 | -0.85 | -2.89 | 1.117 |
| 12 | -6.145 | -6.979 | 1.35 | -1.959 |
| 13 | -5.972 | 1.382 | -4.708 | -5.48 |
| 14 | -6.354 | 1.384 | -5.824 | -5.387 |
| 15 | 1.384 | -5.696 | -6.979 | -5.152 |
| 16 | 1.166 | -4.664 | -0.281 | -3.633 |
| 17 | 0.126 | -0.009 | -0.026 | -0.104 |
| 18 | 0.515 | -0.964 | -0.628 | 0.345 |
| 19 | 0.3 | -0.214 | -0.356 | 0.133 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.85099 |
| 0.0005 | 0.86526 |
| 0.0001 | 4.50501 |