| Motif | NFIA.H12INVIVO.1.PS.A |
| Gene (human) | NFIA (GeneCards) |
| Gene synonyms (human) | KIAA1439 |
| Gene (mouse) | Nfia |
| Gene synonyms (mouse) | |
| LOGO | 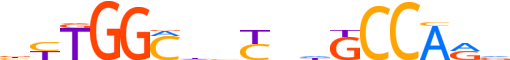 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | A |
| Motif | NFIA.H12INVIVO.1.PS.A |
| Gene (human) | NFIA (GeneCards) |
| Gene synonyms (human) | KIAA1439 |
| Gene (mouse) | Nfia |
| Gene synonyms (mouse) | |
| LOGO | 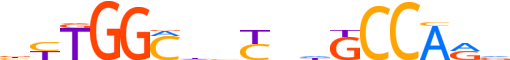 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | A |
| Motif length | 17 |
| Consensus | bYTGGChbCndGCCARv |
| GC content | 62.26% |
| Information content (bits; total / per base) | 15.173 / 0.893 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 999 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (21) | 0.97 | 0.988 | 0.956 | 0.973 | 0.96 | 0.978 | 4.674 | 5.153 | 497.523 | 723.337 |
| Mouse | 6 (28) | 0.812 | 0.876 | 0.711 | 0.812 | 0.76 | 0.839 | 2.248 | 2.933 | 65.94 | 163.886 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 1.0 | 1.0 | 0.986 | 0.987 | 0.819 | 0.854 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.887 | 0.909 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 14.138 | 2.885 | 0.324 | 0.151 |
| TF superclass | beta-Hairpin exposed by an alpha/beta-scaffold {7} (TFClass) |
| TF class | SMAD/NF-1 DNA-binding domain factors {7.1} (TFClass) |
| TF family | NF-1 {7.1.2} (TFClass) |
| TF subfamily | {7.1.2.0} (TFClass) |
| TFClass ID | TFClass: 7.1.2.0.1 |
| HGNC | HGNC:7784 |
| MGI | MGI:108056 |
| EntrezGene (human) | GeneID:4774 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18027 (SSTAR profile) |
| UniProt ID (human) | NFIA_HUMAN |
| UniProt ID (mouse) | NFIA_MOUSE |
| UniProt AC (human) | Q12857 (TFClass) |
| UniProt AC (mouse) | Q02780 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 6 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | NFIA.H12INVIVO.1.PS.A.pcm |
| PWM | NFIA.H12INVIVO.1.PS.A.pwm |
| PFM | NFIA.H12INVIVO.1.PS.A.pfm |
| Alignment | NFIA.H12INVIVO.1.PS.A.words.tsv |
| Threshold to P-value map | NFIA.H12INVIVO.1.PS.A.thr |
| Motif in other formats | |
| JASPAR format | NFIA.H12INVIVO.1.PS.A_jaspar_format.txt |
| MEME format | NFIA.H12INVIVO.1.PS.A_meme_format.meme |
| Transfac format | NFIA.H12INVIVO.1.PS.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 98.0 | 448.0 | 226.0 | 227.0 |
| 02 | 28.0 | 459.0 | 41.0 | 471.0 |
| 03 | 31.0 | 7.0 | 106.0 | 855.0 |
| 04 | 0.0 | 1.0 | 995.0 | 3.0 |
| 05 | 15.0 | 3.0 | 976.0 | 5.0 |
| 06 | 178.0 | 773.0 | 35.0 | 13.0 |
| 07 | 343.0 | 219.0 | 98.0 | 339.0 |
| 08 | 122.0 | 392.0 | 264.0 | 221.0 |
| 09 | 0.0 | 530.0 | 0.0 | 469.0 |
| 10 | 198.0 | 307.0 | 257.0 | 237.0 |
| 11 | 237.0 | 95.0 | 253.0 | 414.0 |
| 12 | 8.0 | 25.0 | 755.0 | 211.0 |
| 13 | 4.0 | 978.0 | 3.0 | 14.0 |
| 14 | 0.0 | 992.0 | 4.0 | 3.0 |
| 15 | 866.0 | 106.0 | 4.0 | 23.0 |
| 16 | 426.0 | 41.0 | 490.0 | 42.0 |
| 17 | 197.0 | 233.0 | 457.0 | 112.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.098 | 0.448 | 0.226 | 0.227 |
| 02 | 0.028 | 0.459 | 0.041 | 0.471 |
| 03 | 0.031 | 0.007 | 0.106 | 0.856 |
| 04 | 0.0 | 0.001 | 0.996 | 0.003 |
| 05 | 0.015 | 0.003 | 0.977 | 0.005 |
| 06 | 0.178 | 0.774 | 0.035 | 0.013 |
| 07 | 0.343 | 0.219 | 0.098 | 0.339 |
| 08 | 0.122 | 0.392 | 0.264 | 0.221 |
| 09 | 0.0 | 0.531 | 0.0 | 0.469 |
| 10 | 0.198 | 0.307 | 0.257 | 0.237 |
| 11 | 0.237 | 0.095 | 0.253 | 0.414 |
| 12 | 0.008 | 0.025 | 0.756 | 0.211 |
| 13 | 0.004 | 0.979 | 0.003 | 0.014 |
| 14 | 0.0 | 0.993 | 0.004 | 0.003 |
| 15 | 0.867 | 0.106 | 0.004 | 0.023 |
| 16 | 0.426 | 0.041 | 0.49 | 0.042 |
| 17 | 0.197 | 0.233 | 0.457 | 0.112 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.925 | 0.581 | -0.099 | -0.095 |
| 02 | -2.135 | 0.605 | -1.773 | 0.631 |
| 03 | -2.039 | -3.361 | -0.848 | 1.226 |
| 04 | -4.981 | -4.524 | 1.377 | -3.974 |
| 05 | -2.71 | -3.974 | 1.358 | -3.621 |
| 06 | -0.336 | 1.125 | -1.924 | -2.838 |
| 07 | 0.315 | -0.13 | -0.925 | 0.304 |
| 08 | -0.709 | 0.448 | 0.055 | -0.121 |
| 09 | -4.981 | 0.749 | -4.981 | 0.627 |
| 10 | -0.23 | 0.205 | 0.028 | -0.052 |
| 11 | -0.052 | -0.955 | 0.013 | 0.503 |
| 12 | -3.252 | -2.242 | 1.102 | -0.167 |
| 13 | -3.782 | 1.36 | -3.974 | -2.772 |
| 14 | -4.981 | 1.374 | -3.782 | -3.974 |
| 15 | 1.239 | -0.848 | -3.782 | -2.319 |
| 16 | 0.531 | -1.773 | 0.671 | -1.749 |
| 17 | -0.235 | -0.069 | 0.601 | -0.794 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.46911 |
| 0.0005 | 3.77266 |
| 0.0001 | 6.47016 |