| Motif | NFAC2.H12CORE.2.S.C |
| Gene (human) | NFATC2 (GeneCards) |
| Gene synonyms (human) | NFAT1, NFATP |
| Gene (mouse) | Nfatc2 |
| Gene synonyms (mouse) | Nfat1, Nfatp |
| LOGO |  |
| LOGO (reverse complement) | 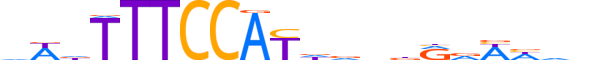 |
| Motif subtype | 2 |
| Quality | C |
| Motif | NFAC2.H12CORE.2.S.C |
| Gene (human) | NFATC2 (GeneCards) |
| Gene synonyms (human) | NFAT1, NFATP |
| Gene (mouse) | Nfatc2 |
| Gene synonyms (mouse) | Nfat1, Nfatp |
| LOGO |  |
| LOGO (reverse complement) | 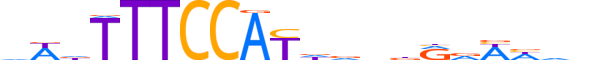 |
| Motif subtype | 2 |
| Quality | C |
| Motif length | 20 |
| Consensus | nbdWhYhnbhATGGAAAhWh |
| GC content | 37.03% |
| Information content (bits; total / per base) | 15.685 / 0.784 |
| Data sources | HT-SELEX |
| Aligned words | 9609 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (10) | 0.74 | 0.767 | 0.585 | 0.617 | 0.72 | 0.733 | 2.028 | 2.132 | 59.845 | 106.328 |
| Mouse | 1 (7) | 0.805 | 0.812 | 0.651 | 0.656 | 0.762 | 0.764 | 2.13 | 2.169 | 95.367 | 106.041 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.992 | 0.988 | 0.982 | 0.974 | 0.944 | 0.932 |
| best | 1.0 | 1.0 | 0.998 | 0.998 | 0.979 | 0.977 | |
| Methyl HT-SELEX, 2 experiments | median | 0.983 | 0.975 | 0.968 | 0.955 | 0.928 | 0.913 |
| best | 1.0 | 1.0 | 0.998 | 0.998 | 0.979 | 0.977 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.992 | 0.988 | 0.982 | 0.974 | 0.944 | 0.932 |
| best | 0.999 | 0.999 | 0.993 | 0.99 | 0.956 | 0.951 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Rel homology region (RHR) factors {6.1} (TFClass) |
| TF family | NFAT-related {6.1.3} (TFClass) |
| TF subfamily | {6.1.3.0} (TFClass) |
| TFClass ID | TFClass: 6.1.3.0.2 |
| HGNC | HGNC:7776 |
| MGI | MGI:102463 |
| EntrezGene (human) | GeneID:4773 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18019 (SSTAR profile) |
| UniProt ID (human) | NFAC2_HUMAN |
| UniProt ID (mouse) | NFAC2_MOUSE |
| UniProt AC (human) | Q13469 (TFClass) |
| UniProt AC (mouse) | Q60591 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 1 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | NFAC2.H12CORE.2.S.C.pcm |
| PWM | NFAC2.H12CORE.2.S.C.pwm |
| PFM | NFAC2.H12CORE.2.S.C.pfm |
| Alignment | NFAC2.H12CORE.2.S.C.words.tsv |
| Threshold to P-value map | NFAC2.H12CORE.2.S.C.thr |
| Motif in other formats | |
| JASPAR format | NFAC2.H12CORE.2.S.C_jaspar_format.txt |
| MEME format | NFAC2.H12CORE.2.S.C_meme_format.meme |
| Transfac format | NFAC2.H12CORE.2.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2334.0 | 1743.0 | 2130.0 | 3402.0 |
| 02 | 1558.25 | 1656.25 | 2079.25 | 4315.25 |
| 03 | 1434.0 | 1180.0 | 1427.0 | 5568.0 |
| 04 | 1301.0 | 843.0 | 845.0 | 6620.0 |
| 05 | 945.0 | 3325.0 | 921.0 | 4418.0 |
| 06 | 831.0 | 6303.0 | 841.0 | 1634.0 |
| 07 | 2653.0 | 4026.0 | 1405.0 | 1525.0 |
| 08 | 3000.0 | 1999.0 | 2026.0 | 2584.0 |
| 09 | 978.0 | 3359.0 | 1461.0 | 3811.0 |
| 10 | 4206.0 | 1638.0 | 924.0 | 2841.0 |
| 11 | 6752.0 | 46.0 | 2771.0 | 40.0 |
| 12 | 1.0 | 558.0 | 0.0 | 9050.0 |
| 13 | 0.0 | 0.0 | 9609.0 | 0.0 |
| 14 | 0.0 | 0.0 | 9609.0 | 0.0 |
| 15 | 9609.0 | 0.0 | 0.0 | 0.0 |
| 16 | 9608.0 | 1.0 | 0.0 | 0.0 |
| 17 | 8847.0 | 283.0 | 14.0 | 465.0 |
| 18 | 3862.0 | 2433.0 | 981.0 | 2333.0 |
| 19 | 2587.75 | 553.75 | 393.75 | 6073.75 |
| 20 | 2106.0 | 2429.0 | 1360.0 | 3714.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.243 | 0.181 | 0.222 | 0.354 |
| 02 | 0.162 | 0.172 | 0.216 | 0.449 |
| 03 | 0.149 | 0.123 | 0.149 | 0.579 |
| 04 | 0.135 | 0.088 | 0.088 | 0.689 |
| 05 | 0.098 | 0.346 | 0.096 | 0.46 |
| 06 | 0.086 | 0.656 | 0.088 | 0.17 |
| 07 | 0.276 | 0.419 | 0.146 | 0.159 |
| 08 | 0.312 | 0.208 | 0.211 | 0.269 |
| 09 | 0.102 | 0.35 | 0.152 | 0.397 |
| 10 | 0.438 | 0.17 | 0.096 | 0.296 |
| 11 | 0.703 | 0.005 | 0.288 | 0.004 |
| 12 | 0.0 | 0.058 | 0.0 | 0.942 |
| 13 | 0.0 | 0.0 | 1.0 | 0.0 |
| 14 | 0.0 | 0.0 | 1.0 | 0.0 |
| 15 | 1.0 | 0.0 | 0.0 | 0.0 |
| 16 | 1.0 | 0.0 | 0.0 | 0.0 |
| 17 | 0.921 | 0.029 | 0.001 | 0.048 |
| 18 | 0.402 | 0.253 | 0.102 | 0.243 |
| 19 | 0.269 | 0.058 | 0.041 | 0.632 |
| 20 | 0.219 | 0.253 | 0.142 | 0.387 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.029 | -0.32 | -0.12 | 0.348 |
| 02 | -0.432 | -0.371 | -0.144 | 0.585 |
| 03 | -0.515 | -0.71 | -0.52 | 0.84 |
| 04 | -0.612 | -1.045 | -1.043 | 1.013 |
| 05 | -0.932 | 0.325 | -0.957 | 0.609 |
| 06 | -1.06 | 0.964 | -1.048 | -0.385 |
| 07 | 0.099 | 0.516 | -0.536 | -0.454 |
| 08 | 0.222 | -0.184 | -0.17 | 0.073 |
| 09 | -0.897 | 0.335 | -0.497 | 0.461 |
| 10 | 0.56 | -0.382 | -0.954 | 0.168 |
| 11 | 1.033 | -3.908 | 0.143 | -4.041 |
| 12 | -6.593 | -1.457 | -6.955 | 1.326 |
| 13 | -6.955 | -6.955 | 1.386 | -6.955 |
| 14 | -6.955 | -6.955 | 1.386 | -6.955 |
| 15 | 1.386 | -6.955 | -6.955 | -6.955 |
| 16 | 1.385 | -6.593 | -6.955 | -6.955 |
| 17 | 1.303 | -2.132 | -4.994 | -1.638 |
| 18 | 0.474 | 0.013 | -0.894 | -0.029 |
| 19 | 0.074 | -1.464 | -1.804 | 0.927 |
| 20 | -0.131 | 0.011 | -0.568 | 0.435 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.63486 |
| 0.0005 | 2.50196 |
| 0.0001 | 6.22296 |