| Motif | MYOD1.H12INVITRO.0.P.B |
| Gene (human) | MYOD1 (GeneCards) |
| Gene synonyms (human) | BHLHC1, MYF3, MYOD |
| Gene (mouse) | Myod1 |
| Gene synonyms (mouse) | Myod |
| LOGO | 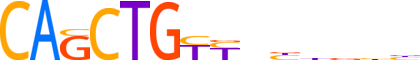 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | MYOD1.H12INVITRO.0.P.B |
| Gene (human) | MYOD1 (GeneCards) |
| Gene synonyms (human) | BHLHC1, MYF3, MYOD |
| Gene (mouse) | Myod1 |
| Gene synonyms (mouse) | Myod |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | CAGCTGYYbbbbbb |
| GC content | 59.33% |
| Information content (bits; total / per base) | 13.191 / 0.942 |
| Data sources | ChIP-Seq |
| Aligned words | 999 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 12 (70) | 0.943 | 0.977 | 0.877 | 0.935 | 0.919 | 0.959 | 3.613 | 4.226 | 298.608 | 614.432 |
| Mouse | 31 (194) | 0.946 | 0.974 | 0.884 | 0.941 | 0.921 | 0.951 | 3.673 | 4.256 | 344.208 | 560.553 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.915 | 0.882 | 0.834 | 0.804 | 0.719 | 0.713 |
| best | 0.954 | 0.934 | 0.888 | 0.859 | 0.777 | 0.762 | |
| Methyl HT-SELEX, 2 experiments | median | 0.84 | 0.796 | 0.742 | 0.711 | 0.644 | 0.639 |
| best | 0.882 | 0.841 | 0.784 | 0.751 | 0.675 | 0.668 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.951 | 0.928 | 0.886 | 0.857 | 0.771 | 0.76 |
| best | 0.954 | 0.934 | 0.888 | 0.859 | 0.777 | 0.762 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 3.448 | 3.677 | 0.362 | 0.172 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.914 | 0.575 | 0.866 | 0.551 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | MyoD-ASC-related {1.2.2} (TFClass) |
| TF subfamily | Myogenic TFs {1.2.2.1} (TFClass) |
| TFClass ID | TFClass: 1.2.2.1.1 |
| HGNC | HGNC:7611 |
| MGI | MGI:97275 |
| EntrezGene (human) | GeneID:4654 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17927 (SSTAR profile) |
| UniProt ID (human) | MYOD1_HUMAN |
| UniProt ID (mouse) | MYOD1_MOUSE |
| UniProt AC (human) | P15172 (TFClass) |
| UniProt AC (mouse) | P10085 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 12 human, 31 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | MYOD1.H12INVITRO.0.P.B.pcm |
| PWM | MYOD1.H12INVITRO.0.P.B.pwm |
| PFM | MYOD1.H12INVITRO.0.P.B.pfm |
| Alignment | MYOD1.H12INVITRO.0.P.B.words.tsv |
| Threshold to P-value map | MYOD1.H12INVITRO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | MYOD1.H12INVITRO.0.P.B_jaspar_format.txt |
| MEME format | MYOD1.H12INVITRO.0.P.B_meme_format.meme |
| Transfac format | MYOD1.H12INVITRO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1.0 | 997.0 | 1.0 | 0.0 |
| 02 | 999.0 | 0.0 | 0.0 | 0.0 |
| 03 | 5.0 | 216.0 | 755.0 | 23.0 |
| 04 | 14.0 | 971.0 | 6.0 | 8.0 |
| 05 | 2.0 | 1.0 | 1.0 | 995.0 |
| 06 | 0.0 | 2.0 | 994.0 | 3.0 |
| 07 | 1.0 | 360.0 | 37.0 | 601.0 |
| 08 | 8.0 | 318.0 | 83.0 | 590.0 |
| 09 | 147.0 | 342.0 | 320.0 | 190.0 |
| 10 | 110.0 | 516.0 | 111.0 | 262.0 |
| 11 | 161.0 | 223.0 | 180.0 | 435.0 |
| 12 | 127.0 | 210.0 | 449.0 | 213.0 |
| 13 | 104.0 | 289.0 | 243.0 | 363.0 |
| 14 | 123.0 | 477.0 | 196.0 | 203.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.001 | 0.998 | 0.001 | 0.0 |
| 02 | 1.0 | 0.0 | 0.0 | 0.0 |
| 03 | 0.005 | 0.216 | 0.756 | 0.023 |
| 04 | 0.014 | 0.972 | 0.006 | 0.008 |
| 05 | 0.002 | 0.001 | 0.001 | 0.996 |
| 06 | 0.0 | 0.002 | 0.995 | 0.003 |
| 07 | 0.001 | 0.36 | 0.037 | 0.602 |
| 08 | 0.008 | 0.318 | 0.083 | 0.591 |
| 09 | 0.147 | 0.342 | 0.32 | 0.19 |
| 10 | 0.11 | 0.517 | 0.111 | 0.262 |
| 11 | 0.161 | 0.223 | 0.18 | 0.435 |
| 12 | 0.127 | 0.21 | 0.449 | 0.213 |
| 13 | 0.104 | 0.289 | 0.243 | 0.363 |
| 14 | 0.123 | 0.477 | 0.196 | 0.203 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -4.524 | 1.379 | -4.524 | -4.981 |
| 02 | 1.381 | -4.981 | -4.981 | -4.981 |
| 03 | -3.621 | -0.144 | 1.102 | -2.319 |
| 04 | -2.772 | 1.353 | -3.483 | -3.252 |
| 05 | -4.212 | -4.524 | -4.524 | 1.377 |
| 06 | -4.981 | -4.212 | 1.376 | -3.974 |
| 07 | -4.524 | 0.364 | -1.871 | 0.874 |
| 08 | -3.252 | 0.24 | -1.088 | 0.856 |
| 09 | -0.525 | 0.312 | 0.246 | -0.271 |
| 10 | -0.811 | 0.722 | -0.802 | 0.048 |
| 11 | -0.435 | -0.112 | -0.325 | 0.552 |
| 12 | -0.67 | -0.172 | 0.584 | -0.158 |
| 13 | -0.866 | 0.145 | -0.027 | 0.372 |
| 14 | -0.701 | 0.644 | -0.24 | -0.206 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.13916 |
| 0.0005 | 4.30721 |
| 0.0001 | 7.295265 |