| Motif | MITF.H12INVIVO.0.P.B |
| Gene (human) | MITF (GeneCards) |
| Gene synonyms (human) | BHLHE32 |
| Gene (mouse) | Mitf |
| Gene synonyms (mouse) | Bw, Mi, Vit |
| LOGO |  |
| LOGO (reverse complement) | 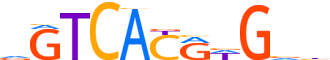 |
| Motif subtype | 0 |
| Quality | B |
| Motif | MITF.H12INVIVO.0.P.B |
| Gene (human) | MITF (GeneCards) |
| Gene synonyms (human) | BHLHE32 |
| Gene (mouse) | Mitf |
| Gene synonyms (mouse) | Bw, Mi, Vit |
| LOGO |  |
| LOGO (reverse complement) | 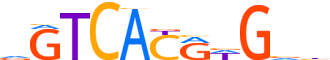 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 11 |
| Consensus | vbChYGTGACh |
| GC content | 57.23% |
| Information content (bits; total / per base) | 11.198 / 1.018 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 10 (58) | 0.887 | 0.996 | 0.807 | 0.987 | 0.856 | 0.997 | 3.558 | 7.97 | 189.136 | 1120.62 |
| Mouse | 1 (5) | 0.821 | 0.832 | 0.723 | 0.738 | 0.777 | 0.793 | 2.584 | 2.649 | 177.222 | 191.886 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 17.184 | 12.587 | 0.287 | 0.235 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | bHLH-ZIP {1.2.6} (TFClass) |
| TF subfamily | TFE3 {1.2.6.1} (TFClass) |
| TFClass ID | TFClass: 1.2.6.1.4 |
| HGNC | HGNC:7105 |
| MGI | MGI:104554 |
| EntrezGene (human) | GeneID:4286 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17342 (SSTAR profile) |
| UniProt ID (human) | MITF_HUMAN |
| UniProt ID (mouse) | MITF_MOUSE |
| UniProt AC (human) | O75030 (TFClass) |
| UniProt AC (mouse) | Q08874 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 10 human, 1 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | MITF.H12INVIVO.0.P.B.pcm |
| PWM | MITF.H12INVIVO.0.P.B.pwm |
| PFM | MITF.H12INVIVO.0.P.B.pfm |
| Alignment | MITF.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | MITF.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | MITF.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | MITF.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | MITF.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 330.0 | 187.0 | 366.0 | 117.0 |
| 02 | 74.0 | 261.0 | 255.0 | 410.0 |
| 03 | 10.0 | 983.0 | 4.0 | 3.0 |
| 04 | 578.0 | 104.0 | 88.0 | 230.0 |
| 05 | 5.0 | 674.0 | 65.0 | 256.0 |
| 06 | 139.0 | 72.0 | 788.0 | 1.0 |
| 07 | 7.0 | 1.0 | 28.0 | 964.0 |
| 08 | 0.0 | 3.0 | 995.0 | 2.0 |
| 09 | 953.0 | 18.0 | 15.0 | 14.0 |
| 10 | 5.0 | 845.0 | 34.0 | 116.0 |
| 11 | 156.0 | 402.0 | 107.0 | 335.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.33 | 0.187 | 0.366 | 0.117 |
| 02 | 0.074 | 0.261 | 0.255 | 0.41 |
| 03 | 0.01 | 0.983 | 0.004 | 0.003 |
| 04 | 0.578 | 0.104 | 0.088 | 0.23 |
| 05 | 0.005 | 0.674 | 0.065 | 0.256 |
| 06 | 0.139 | 0.072 | 0.788 | 0.001 |
| 07 | 0.007 | 0.001 | 0.028 | 0.964 |
| 08 | 0.0 | 0.003 | 0.995 | 0.002 |
| 09 | 0.953 | 0.018 | 0.015 | 0.014 |
| 10 | 0.005 | 0.845 | 0.034 | 0.116 |
| 11 | 0.156 | 0.402 | 0.107 | 0.335 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.276 | -0.288 | 0.379 | -0.752 |
| 02 | -1.201 | 0.043 | 0.02 | 0.492 |
| 03 | -3.066 | 1.364 | -3.783 | -3.975 |
| 04 | 0.834 | -0.867 | -1.032 | -0.083 |
| 05 | -3.622 | 0.987 | -1.328 | 0.024 |
| 06 | -0.582 | -1.228 | 1.143 | -4.525 |
| 07 | -3.362 | -4.525 | -2.136 | 1.345 |
| 08 | -4.982 | -3.975 | 1.376 | -4.213 |
| 09 | 1.333 | -2.546 | -2.711 | -2.773 |
| 10 | -3.622 | 1.213 | -1.952 | -0.76 |
| 11 | -0.467 | 0.472 | -0.84 | 0.291 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.34378 |
| 0.0005 | 5.383285 |
| 0.0001 | 7.47678 |