| Motif | MEIS2.H12CORE.2.SM.B |
| Gene (human) | MEIS2 (GeneCards) |
| Gene synonyms (human) | MRG1 |
| Gene (mouse) | Meis2 |
| Gene synonyms (mouse) | Mrg1, Stra10 |
| LOGO | 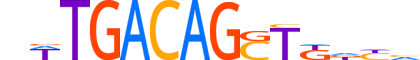 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif | MEIS2.H12CORE.2.SM.B |
| Gene (human) | MEIS2 (GeneCards) |
| Gene synonyms (human) | MRG1 |
| Gene (mouse) | Meis2 |
| Gene synonyms (mouse) | Mrg1, Stra10 |
| LOGO | 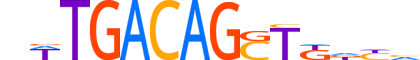 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 14 |
| Consensus | nWTGACAGSTKdhv |
| GC content | 48.39% |
| Information content (bits; total / per base) | 16.129 / 1.152 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 616 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.679 | 0.69 | 0.542 | 0.561 | 0.611 | 0.639 | 1.489 | 1.632 | 72.796 | 105.387 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 8 experiments | median | 0.964 | 0.948 | 0.905 | 0.882 | 0.794 | 0.783 |
| best | 0.997 | 0.996 | 0.993 | 0.989 | 0.985 | 0.978 | |
| Methyl HT-SELEX, 2 experiments | median | 0.964 | 0.948 | 0.905 | 0.882 | 0.794 | 0.783 |
| best | 0.973 | 0.96 | 0.921 | 0.902 | 0.795 | 0.792 | |
| Non-Methyl HT-SELEX, 6 experiments | median | 0.911 | 0.886 | 0.842 | 0.817 | 0.741 | 0.732 |
| best | 0.997 | 0.996 | 0.993 | 0.989 | 0.985 | 0.978 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 5.732 | 3.646 | 0.181 | 0.183 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.685 | 0.333 | 0.405 | 0.177 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | TALE-type HD {3.1.4} (TFClass) |
| TF subfamily | MEIS {3.1.4.2} (TFClass) |
| TFClass ID | TFClass: 3.1.4.2.2 |
| HGNC | HGNC:7001 |
| MGI | MGI:108564 |
| EntrezGene (human) | GeneID:4212 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17536 (SSTAR profile) |
| UniProt ID (human) | MEIS2_HUMAN |
| UniProt ID (mouse) | MEIS2_MOUSE |
| UniProt AC (human) | O14770 (TFClass) |
| UniProt AC (mouse) | P97367 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 6 |
| Methyl-HT-SELEX | 2 |
| PCM | MEIS2.H12CORE.2.SM.B.pcm |
| PWM | MEIS2.H12CORE.2.SM.B.pwm |
| PFM | MEIS2.H12CORE.2.SM.B.pfm |
| Alignment | MEIS2.H12CORE.2.SM.B.words.tsv |
| Threshold to P-value map | MEIS2.H12CORE.2.SM.B.thr |
| Motif in other formats | |
| JASPAR format | MEIS2.H12CORE.2.SM.B_jaspar_format.txt |
| MEME format | MEIS2.H12CORE.2.SM.B_meme_format.meme |
| Transfac format | MEIS2.H12CORE.2.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 148.25 | 148.25 | 132.25 | 187.25 |
| 02 | 112.5 | 52.5 | 50.5 | 400.5 |
| 03 | 0.0 | 0.0 | 0.0 | 616.0 |
| 04 | 0.0 | 0.0 | 616.0 | 0.0 |
| 05 | 616.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 616.0 | 0.0 | 0.0 |
| 07 | 616.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 613.0 | 3.0 |
| 09 | 7.0 | 395.0 | 212.0 | 2.0 |
| 10 | 6.0 | 73.0 | 14.0 | 523.0 |
| 11 | 45.0 | 32.0 | 372.0 | 167.0 |
| 12 | 132.0 | 45.0 | 152.0 | 287.0 |
| 13 | 78.5 | 353.5 | 51.5 | 132.5 |
| 14 | 295.25 | 167.25 | 77.25 | 76.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.241 | 0.241 | 0.215 | 0.304 |
| 02 | 0.183 | 0.085 | 0.082 | 0.65 |
| 03 | 0.0 | 0.0 | 0.0 | 1.0 |
| 04 | 0.0 | 0.0 | 1.0 | 0.0 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 1.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.995 | 0.005 |
| 09 | 0.011 | 0.641 | 0.344 | 0.003 |
| 10 | 0.01 | 0.119 | 0.023 | 0.849 |
| 11 | 0.073 | 0.052 | 0.604 | 0.271 |
| 12 | 0.214 | 0.073 | 0.247 | 0.466 |
| 13 | 0.127 | 0.574 | 0.084 | 0.215 |
| 14 | 0.479 | 0.272 | 0.125 | 0.124 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.038 | -0.038 | -0.151 | 0.194 |
| 02 | -0.31 | -1.056 | -1.094 | 0.949 |
| 03 | -4.574 | -4.574 | -4.574 | 1.379 |
| 04 | -4.574 | -4.574 | 1.379 | -4.574 |
| 05 | 1.379 | -4.574 | -4.574 | -4.574 |
| 06 | -4.574 | 1.379 | -4.574 | -4.574 |
| 07 | 1.379 | -4.574 | -4.574 | -4.574 |
| 08 | -4.574 | -4.574 | 1.374 | -3.52 |
| 09 | -2.895 | 0.936 | 0.317 | -3.765 |
| 10 | -3.018 | -0.735 | -2.3 | 1.215 |
| 11 | -1.206 | -1.533 | 0.876 | 0.08 |
| 12 | -0.152 | -1.206 | -0.013 | 0.618 |
| 13 | -0.664 | 0.825 | -1.075 | -0.149 |
| 14 | 0.646 | 0.082 | -0.68 | -0.692 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.37952 |
| 0.0005 | 2.91001 |
| 0.0001 | 5.85332 |