| Motif | MEF2B.H12CORE.1.SM.B |
| Gene (human) | MEF2B (GeneCards) |
| Gene synonyms (human) | XMEF2 |
| Gene (mouse) | Mef2b |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 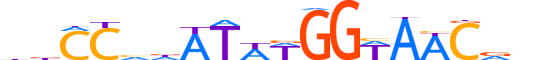 |
| Motif subtype | 1 |
| Quality | B |
| Motif | MEF2B.H12CORE.1.SM.B |
| Gene (human) | MEF2B (GeneCards) |
| Gene synonyms (human) | XMEF2 |
| Gene (mouse) | Mef2b |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | nhGWTWCCWWAThbGGhh |
| GC content | 42.12% |
| Information content (bits; total / per base) | 15.516 / 0.862 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 7295 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 16 (100) | 0.696 | 0.761 | 0.532 | 0.611 | 0.623 | 0.692 | 1.578 | 1.904 | 50.947 | 144.229 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 3 experiments | median | 0.98 | 0.974 | 0.91 | 0.896 | 0.787 | 0.785 |
| best | 0.999 | 0.998 | 0.988 | 0.985 | 0.918 | 0.914 | |
| Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.998 | 0.988 | 0.985 | 0.918 | 0.914 |
| best | 0.999 | 0.998 | 0.988 | 0.985 | 0.918 | 0.914 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.753 | 0.752 | 0.706 | 0.704 | 0.641 | 0.645 |
| best | 0.98 | 0.974 | 0.91 | 0.896 | 0.787 | 0.785 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.855 | 1.938 | 0.103 | 0.068 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.799 | 0.338 | 0.43 | 0.24 |
| batch 2 | 0.778 | 0.365 | 0.585 | 0.467 |
| TF superclass | alpha-Helices exposed by beta-structures {5} (TFClass) |
| TF class | MADS box factors {5.1} (TFClass) |
| TF family | Regulators of differentiation {5.1.1} (TFClass) |
| TF subfamily | MEF2 {5.1.1.1} (TFClass) |
| TFClass ID | TFClass: 5.1.1.1.2 |
| HGNC | HGNC:6995 |
| MGI | MGI:104526 |
| EntrezGene (human) | GeneID:100271849 (SSTAR profile) GeneID:4207 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | MEF2B_HUMAN |
| UniProt ID (mouse) | MEF2B_MOUSE |
| UniProt AC (human) | Q02080 (TFClass) |
| UniProt AC (mouse) | O55087 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 16 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 1 |
| PCM | MEF2B.H12CORE.1.SM.B.pcm |
| PWM | MEF2B.H12CORE.1.SM.B.pwm |
| PFM | MEF2B.H12CORE.1.SM.B.pfm |
| Alignment | MEF2B.H12CORE.1.SM.B.words.tsv |
| Threshold to P-value map | MEF2B.H12CORE.1.SM.B.thr |
| Motif in other formats | |
| JASPAR format | MEF2B.H12CORE.1.SM.B_jaspar_format.txt |
| MEME format | MEF2B.H12CORE.1.SM.B_meme_format.meme |
| Transfac format | MEF2B.H12CORE.1.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1829.5 | 1601.5 | 2325.5 | 1538.5 |
| 02 | 716.0 | 2990.0 | 553.0 | 3036.0 |
| 03 | 218.0 | 362.0 | 6537.0 | 178.0 |
| 04 | 807.0 | 232.0 | 441.0 | 5815.0 |
| 05 | 362.0 | 91.0 | 142.0 | 6700.0 |
| 06 | 5036.0 | 132.0 | 864.0 | 1263.0 |
| 07 | 1.0 | 7293.0 | 0.0 | 1.0 |
| 08 | 1.0 | 6993.0 | 2.0 | 299.0 |
| 09 | 5079.0 | 582.0 | 443.0 | 1191.0 |
| 10 | 2419.0 | 334.0 | 311.0 | 4231.0 |
| 11 | 6175.0 | 11.0 | 27.0 | 1082.0 |
| 12 | 1433.0 | 126.0 | 41.0 | 5695.0 |
| 13 | 2197.0 | 1178.0 | 683.0 | 3237.0 |
| 14 | 903.0 | 1050.0 | 1920.0 | 3422.0 |
| 15 | 1189.0 | 16.0 | 6042.0 | 48.0 |
| 16 | 242.0 | 128.0 | 6087.0 | 838.0 |
| 17 | 2770.75 | 2405.75 | 837.75 | 1280.75 |
| 18 | 3067.5 | 1390.5 | 1130.5 | 1706.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.251 | 0.22 | 0.319 | 0.211 |
| 02 | 0.098 | 0.41 | 0.076 | 0.416 |
| 03 | 0.03 | 0.05 | 0.896 | 0.024 |
| 04 | 0.111 | 0.032 | 0.06 | 0.797 |
| 05 | 0.05 | 0.012 | 0.019 | 0.918 |
| 06 | 0.69 | 0.018 | 0.118 | 0.173 |
| 07 | 0.0 | 1.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.959 | 0.0 | 0.041 |
| 09 | 0.696 | 0.08 | 0.061 | 0.163 |
| 10 | 0.332 | 0.046 | 0.043 | 0.58 |
| 11 | 0.846 | 0.002 | 0.004 | 0.148 |
| 12 | 0.196 | 0.017 | 0.006 | 0.781 |
| 13 | 0.301 | 0.161 | 0.094 | 0.444 |
| 14 | 0.124 | 0.144 | 0.263 | 0.469 |
| 15 | 0.163 | 0.002 | 0.828 | 0.007 |
| 16 | 0.033 | 0.018 | 0.834 | 0.115 |
| 17 | 0.38 | 0.33 | 0.115 | 0.176 |
| 18 | 0.42 | 0.191 | 0.155 | 0.234 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.003 | -0.13 | 0.243 | -0.17 |
| 02 | -0.933 | 0.494 | -1.19 | 0.509 |
| 03 | -2.115 | -1.612 | 1.276 | -2.316 |
| 04 | -0.814 | -2.054 | -1.416 | 1.159 |
| 05 | -1.612 | -2.975 | -2.539 | 1.3 |
| 06 | 1.015 | -2.61 | -0.746 | -0.367 |
| 07 | -6.339 | 1.385 | -6.711 | -6.339 |
| 08 | -6.339 | 1.343 | -6.069 | -1.802 |
| 09 | 1.023 | -1.14 | -1.411 | -0.425 |
| 10 | 0.282 | -1.692 | -1.763 | 0.841 |
| 11 | 1.219 | -4.928 | -4.135 | -0.521 |
| 12 | -0.241 | -2.656 | -3.743 | 1.138 |
| 13 | 0.186 | -0.436 | -0.98 | 0.573 |
| 14 | -0.702 | -0.551 | 0.051 | 0.629 |
| 15 | -0.427 | -4.607 | 1.197 | -3.593 |
| 16 | -2.012 | -2.641 | 1.204 | -0.776 |
| 17 | 0.418 | 0.277 | -0.776 | -0.353 |
| 18 | 0.519 | -0.271 | -0.477 | -0.066 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.57061 |
| 0.0005 | 3.83511 |
| 0.0001 | 6.41411 |