| Motif | KLF11.H12CORE.1.SM.B |
| Gene (human) | KLF11 (GeneCards) |
| Gene synonyms (human) | FKLF, TIEG2 |
| Gene (mouse) | Klf11 |
| Gene synonyms (mouse) | Tieg2b, Tieg3 |
| LOGO |  |
| LOGO (reverse complement) | 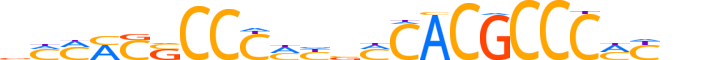 |
| Motif subtype | 1 |
| Quality | B |
| Motif | KLF11.H12CORE.1.SM.B |
| Gene (human) | KLF11 (GeneCards) |
| Gene synonyms (human) | FKLF, TIEG2 |
| Gene (mouse) | Klf11 |
| Gene synonyms (mouse) | Tieg2b, Tieg3 |
| LOGO |  |
| LOGO (reverse complement) | 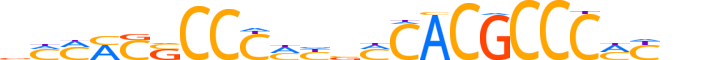 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 24 |
| Consensus | nnRKGGGCGTGKbRKGGGSSKKKb |
| GC content | 73.04% |
| Information content (bits; total / per base) | 23.575 / 0.982 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 7766 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (14) | 0.657 | 0.678 | 0.456 | 0.497 | 0.746 | 0.757 | 2.435 | 2.66 | 4.855 | 8.0 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.995 | 0.992 | 0.989 | 0.983 | 0.955 | 0.947 |
| best | 0.996 | 0.993 | 0.991 | 0.986 | 0.956 | 0.948 | |
| Methyl HT-SELEX, 1 experiments | median | 0.994 | 0.99 | 0.987 | 0.98 | 0.956 | 0.945 |
| best | 0.994 | 0.99 | 0.987 | 0.98 | 0.956 | 0.945 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.996 | 0.993 | 0.991 | 0.986 | 0.955 | 0.948 |
| best | 0.996 | 0.993 | 0.991 | 0.986 | 0.955 | 0.948 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.865 | 0.697 | 0.902 | 0.723 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Three-zinc finger Kruppel-related {2.3.1} (TFClass) |
| TF subfamily | Kr-like {2.3.1.2} (TFClass) |
| TFClass ID | TFClass: 2.3.1.2.11 |
| HGNC | HGNC:11811 |
| MGI | MGI:2653368 |
| EntrezGene (human) | GeneID:8462 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:194655 (SSTAR profile) |
| UniProt ID (human) | KLF11_HUMAN |
| UniProt ID (mouse) | KLF11_MOUSE |
| UniProt AC (human) | O14901 (TFClass) |
| UniProt AC (mouse) | Q8K1S5 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | KLF11.H12CORE.1.SM.B.pcm |
| PWM | KLF11.H12CORE.1.SM.B.pwm |
| PFM | KLF11.H12CORE.1.SM.B.pfm |
| Alignment | KLF11.H12CORE.1.SM.B.words.tsv |
| Threshold to P-value map | KLF11.H12CORE.1.SM.B.thr |
| Motif in other formats | |
| JASPAR format | KLF11.H12CORE.1.SM.B_jaspar_format.txt |
| MEME format | KLF11.H12CORE.1.SM.B_meme_format.meme |
| Transfac format | KLF11.H12CORE.1.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2082.25 | 1053.25 | 2843.25 | 1787.25 |
| 02 | 2226.75 | 1213.75 | 2579.75 | 1745.75 |
| 03 | 1589.0 | 285.0 | 5350.0 | 542.0 |
| 04 | 794.0 | 44.0 | 3632.0 | 3296.0 |
| 05 | 483.0 | 6.0 | 7261.0 | 16.0 |
| 06 | 0.0 | 3.0 | 7761.0 | 2.0 |
| 07 | 1.0 | 0.0 | 7764.0 | 1.0 |
| 08 | 119.0 | 7287.0 | 8.0 | 352.0 |
| 09 | 33.0 | 1.0 | 7704.0 | 28.0 |
| 10 | 23.0 | 29.0 | 477.0 | 7237.0 |
| 11 | 329.0 | 94.0 | 6885.0 | 458.0 |
| 12 | 821.0 | 229.0 | 4920.0 | 1796.0 |
| 13 | 1078.0 | 3717.0 | 1704.0 | 1267.0 |
| 14 | 1640.0 | 673.0 | 4629.0 | 824.0 |
| 15 | 965.0 | 111.0 | 3491.0 | 3199.0 |
| 16 | 780.0 | 36.0 | 6256.0 | 694.0 |
| 17 | 148.0 | 25.0 | 7507.0 | 86.0 |
| 18 | 49.0 | 47.0 | 7603.0 | 67.0 |
| 19 | 321.0 | 4850.0 | 2005.0 | 590.0 |
| 20 | 178.0 | 1602.0 | 5700.0 | 286.0 |
| 21 | 102.0 | 198.0 | 2752.0 | 4714.0 |
| 22 | 441.0 | 207.0 | 4960.0 | 2158.0 |
| 23 | 760.5 | 458.5 | 5021.5 | 1525.5 |
| 24 | 815.75 | 2337.75 | 2817.75 | 1794.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.268 | 0.136 | 0.366 | 0.23 |
| 02 | 0.287 | 0.156 | 0.332 | 0.225 |
| 03 | 0.205 | 0.037 | 0.689 | 0.07 |
| 04 | 0.102 | 0.006 | 0.468 | 0.424 |
| 05 | 0.062 | 0.001 | 0.935 | 0.002 |
| 06 | 0.0 | 0.0 | 0.999 | 0.0 |
| 07 | 0.0 | 0.0 | 1.0 | 0.0 |
| 08 | 0.015 | 0.938 | 0.001 | 0.045 |
| 09 | 0.004 | 0.0 | 0.992 | 0.004 |
| 10 | 0.003 | 0.004 | 0.061 | 0.932 |
| 11 | 0.042 | 0.012 | 0.887 | 0.059 |
| 12 | 0.106 | 0.029 | 0.634 | 0.231 |
| 13 | 0.139 | 0.479 | 0.219 | 0.163 |
| 14 | 0.211 | 0.087 | 0.596 | 0.106 |
| 15 | 0.124 | 0.014 | 0.45 | 0.412 |
| 16 | 0.1 | 0.005 | 0.806 | 0.089 |
| 17 | 0.019 | 0.003 | 0.967 | 0.011 |
| 18 | 0.006 | 0.006 | 0.979 | 0.009 |
| 19 | 0.041 | 0.625 | 0.258 | 0.076 |
| 20 | 0.023 | 0.206 | 0.734 | 0.037 |
| 21 | 0.013 | 0.025 | 0.354 | 0.607 |
| 22 | 0.057 | 0.027 | 0.639 | 0.278 |
| 23 | 0.098 | 0.059 | 0.647 | 0.196 |
| 24 | 0.105 | 0.301 | 0.363 | 0.231 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.07 | -0.611 | 0.381 | -0.083 |
| 02 | 0.137 | -0.469 | 0.284 | -0.106 |
| 03 | -0.2 | -1.912 | 1.013 | -1.273 |
| 04 | -0.892 | -3.739 | 0.626 | 0.529 |
| 05 | -1.388 | -5.463 | 1.318 | -4.669 |
| 06 | -6.766 | -5.916 | 1.385 | -6.128 |
| 07 | -6.397 | -6.766 | 1.385 | -6.397 |
| 08 | -2.775 | 1.322 | -5.246 | -1.702 |
| 09 | -4.01 | -6.397 | 1.377 | -4.163 |
| 10 | -4.344 | -4.131 | -1.4 | 1.315 |
| 11 | -1.77 | -3.006 | 1.265 | -1.441 |
| 12 | -0.859 | -2.129 | 0.929 | -0.078 |
| 13 | -0.587 | 0.649 | -0.13 | -0.426 |
| 14 | -0.169 | -1.057 | 0.868 | -0.855 |
| 15 | -0.698 | -2.843 | 0.586 | 0.499 |
| 16 | -0.91 | -3.929 | 1.169 | -1.027 |
| 17 | -2.56 | -4.268 | 1.352 | -3.092 |
| 18 | -3.636 | -3.676 | 1.364 | -3.335 |
| 19 | -1.794 | 0.915 | 0.032 | -1.188 |
| 20 | -2.378 | -0.192 | 1.076 | -1.909 |
| 21 | -2.926 | -2.273 | 0.349 | 0.886 |
| 22 | -1.478 | -2.229 | 0.937 | 0.106 |
| 23 | -0.935 | -1.44 | 0.95 | -0.241 |
| 24 | -0.866 | 0.186 | 0.372 | -0.079 |
| P-value | Threshold |
|---|---|
| 0.001 | -3.42854 |
| 0.0005 | -1.61149 |
| 0.0001 | 2.20936 |