| Motif | KLF10.H12INVITRO.1.PSM.A |
| Gene (human) | KLF10 (GeneCards) |
| Gene synonyms (human) | TIEG, TIEG1 |
| Gene (mouse) | Klf10 |
| Gene synonyms (mouse) | Gdnfif, Tieg, Tieg1 |
| LOGO |  |
| LOGO (reverse complement) | 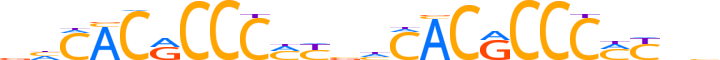 |
| Motif subtype | 1 |
| Quality | A |
| Motif | KLF10.H12INVITRO.1.PSM.A |
| Gene (human) | KLF10 (GeneCards) |
| Gene synonyms (human) | TIEG, TIEG1 |
| Gene (mouse) | Klf10 |
| Gene synonyms (mouse) | Gdnfif, Tieg, Tieg1 |
| LOGO |  |
| LOGO (reverse complement) | 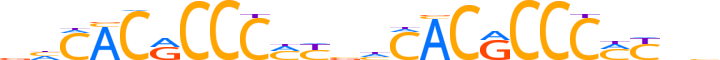 |
| Motif subtype | 1 |
| Quality | A |
| Motif length | 24 |
| Consensus | ddRKGGGYGTGdvRKGGGYGTGdb |
| GC content | 69.78% |
| Information content (bits; total / per base) | 24.956 / 1.04 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 7817 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.732 | 0.796 | 0.613 | 0.716 | 0.782 | 0.857 | 3.348 | 4.31 | 25.357 | 30.538 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.999 | 0.999 | 0.992 | 0.99 | 0.847 | 0.876 |
| best | 0.999 | 0.999 | 0.998 | 0.997 | 0.926 | 0.932 | |
| Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.999 | 0.998 | 0.997 | 0.926 | 0.932 |
| best | 0.999 | 0.999 | 0.998 | 0.997 | 0.926 | 0.932 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.998 | 0.986 | 0.984 | 0.768 | 0.82 |
| best | 0.999 | 0.998 | 0.986 | 0.984 | 0.768 | 0.82 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.824 | 0.267 | 0.821 | 0.643 |
| batch 2 | 0.622 | 0.267 | 0.641 | 0.353 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Three-zinc finger Kruppel-related {2.3.1} (TFClass) |
| TF subfamily | Kr-like {2.3.1.2} (TFClass) |
| TFClass ID | TFClass: 2.3.1.2.10 |
| HGNC | HGNC:11810 |
| MGI | MGI:1101353 |
| EntrezGene (human) | GeneID:7071 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21847 (SSTAR profile) |
| UniProt ID (human) | KLF10_HUMAN |
| UniProt ID (mouse) | KLF10_MOUSE |
| UniProt AC (human) | Q13118 (TFClass) |
| UniProt AC (mouse) | O89091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | KLF10.H12INVITRO.1.PSM.A.pcm |
| PWM | KLF10.H12INVITRO.1.PSM.A.pwm |
| PFM | KLF10.H12INVITRO.1.PSM.A.pfm |
| Alignment | KLF10.H12INVITRO.1.PSM.A.words.tsv |
| Threshold to P-value map | KLF10.H12INVITRO.1.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | KLF10.H12INVITRO.1.PSM.A_jaspar_format.txt |
| MEME format | KLF10.H12INVITRO.1.PSM.A_meme_format.meme |
| Transfac format | KLF10.H12INVITRO.1.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2262.75 | 1004.75 | 2979.75 | 1569.75 |
| 02 | 2448.25 | 1062.25 | 2706.25 | 1600.25 |
| 03 | 1913.0 | 188.0 | 5364.0 | 352.0 |
| 04 | 1120.0 | 49.0 | 4687.0 | 1961.0 |
| 05 | 708.0 | 4.0 | 7095.0 | 10.0 |
| 06 | 2.0 | 3.0 | 7795.0 | 17.0 |
| 07 | 0.0 | 0.0 | 7816.0 | 1.0 |
| 08 | 92.0 | 5227.0 | 16.0 | 2482.0 |
| 09 | 47.0 | 1.0 | 7699.0 | 70.0 |
| 10 | 13.0 | 202.0 | 449.0 | 7153.0 |
| 11 | 391.0 | 220.0 | 6547.0 | 659.0 |
| 12 | 1715.0 | 355.0 | 3247.0 | 2500.0 |
| 13 | 1447.0 | 3209.0 | 2295.0 | 866.0 |
| 14 | 2467.0 | 317.0 | 4557.0 | 476.0 |
| 15 | 1067.0 | 73.0 | 4388.0 | 2289.0 |
| 16 | 695.0 | 4.0 | 7111.0 | 7.0 |
| 17 | 2.0 | 1.0 | 7809.0 | 5.0 |
| 18 | 5.0 | 7.0 | 7801.0 | 4.0 |
| 19 | 104.0 | 4405.0 | 233.0 | 3075.0 |
| 20 | 74.0 | 60.0 | 7494.0 | 189.0 |
| 21 | 27.0 | 260.0 | 847.0 | 6683.0 |
| 22 | 463.0 | 154.0 | 6425.0 | 775.0 |
| 23 | 801.0 | 539.0 | 3110.0 | 3367.0 |
| 24 | 826.5 | 3072.5 | 2027.5 | 1890.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.289 | 0.129 | 0.381 | 0.201 |
| 02 | 0.313 | 0.136 | 0.346 | 0.205 |
| 03 | 0.245 | 0.024 | 0.686 | 0.045 |
| 04 | 0.143 | 0.006 | 0.6 | 0.251 |
| 05 | 0.091 | 0.001 | 0.908 | 0.001 |
| 06 | 0.0 | 0.0 | 0.997 | 0.002 |
| 07 | 0.0 | 0.0 | 1.0 | 0.0 |
| 08 | 0.012 | 0.669 | 0.002 | 0.318 |
| 09 | 0.006 | 0.0 | 0.985 | 0.009 |
| 10 | 0.002 | 0.026 | 0.057 | 0.915 |
| 11 | 0.05 | 0.028 | 0.838 | 0.084 |
| 12 | 0.219 | 0.045 | 0.415 | 0.32 |
| 13 | 0.185 | 0.411 | 0.294 | 0.111 |
| 14 | 0.316 | 0.041 | 0.583 | 0.061 |
| 15 | 0.136 | 0.009 | 0.561 | 0.293 |
| 16 | 0.089 | 0.001 | 0.91 | 0.001 |
| 17 | 0.0 | 0.0 | 0.999 | 0.001 |
| 18 | 0.001 | 0.001 | 0.998 | 0.001 |
| 19 | 0.013 | 0.564 | 0.03 | 0.393 |
| 20 | 0.009 | 0.008 | 0.959 | 0.024 |
| 21 | 0.003 | 0.033 | 0.108 | 0.855 |
| 22 | 0.059 | 0.02 | 0.822 | 0.099 |
| 23 | 0.102 | 0.069 | 0.398 | 0.431 |
| 24 | 0.106 | 0.393 | 0.259 | 0.242 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.146 | -0.664 | 0.421 | -0.219 |
| 02 | 0.225 | -0.609 | 0.325 | -0.2 |
| 03 | -0.021 | -2.331 | 1.009 | -1.709 |
| 04 | -0.556 | -3.642 | 0.874 | 0.003 |
| 05 | -1.013 | -5.748 | 1.289 | -5.074 |
| 06 | -6.134 | -5.922 | 1.383 | -4.622 |
| 07 | -6.772 | -6.772 | 1.385 | -6.403 |
| 08 | -3.033 | 0.983 | -4.675 | 0.239 |
| 09 | -3.682 | -6.403 | 1.37 | -3.299 |
| 10 | -4.855 | -2.26 | -1.467 | 1.297 |
| 11 | -1.604 | -2.175 | 1.208 | -1.085 |
| 12 | -0.13 | -1.7 | 0.507 | 0.246 |
| 13 | -0.3 | 0.496 | 0.161 | -0.812 |
| 14 | 0.233 | -1.813 | 0.846 | -1.409 |
| 15 | -0.604 | -3.258 | 0.808 | 0.158 |
| 16 | -1.032 | -5.748 | 1.291 | -5.355 |
| 17 | -6.134 | -6.403 | 1.384 | -5.599 |
| 18 | -5.599 | -5.355 | 1.383 | -5.748 |
| 19 | -2.913 | 0.812 | -2.118 | 0.453 |
| 20 | -3.245 | -3.448 | 1.343 | -2.325 |
| 21 | -4.203 | -2.01 | -0.835 | 1.229 |
| 22 | -1.436 | -2.528 | 1.189 | -0.923 |
| 23 | -0.89 | -1.285 | 0.464 | 0.544 |
| 24 | -0.859 | 0.452 | 0.037 | -0.033 |
| P-value | Threshold |
|---|---|
| 0.001 | -5.87014 |
| 0.0005 | -3.83569 |
| 0.0001 | 0.45811 |