| Motif | IRF9.H12INVIVO.0.PSM.A |
| Gene (human) | IRF9 (GeneCards) |
| Gene synonyms (human) | ISGF3G |
| Gene (mouse) | Irf9 |
| Gene synonyms (mouse) | Isgf3g |
| LOGO |  |
| LOGO (reverse complement) | 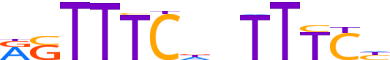 |
| Motif subtype | 0 |
| Quality | A |
| Motif | IRF9.H12INVIVO.0.PSM.A |
| Gene (human) | IRF9 (GeneCards) |
| Gene synonyms (human) | ISGF3G |
| Gene (mouse) | Irf9 |
| Gene synonyms (mouse) | Isgf3g |
| LOGO |  |
| LOGO (reverse complement) | 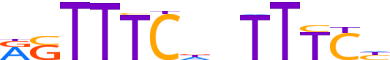 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 13 |
| Consensus | dGAAAnhGAAASY |
| GC content | 36.05% |
| Information content (bits; total / per base) | 15.069 / 1.159 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 994 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 3 (15) | 0.9 | 0.958 | 0.857 | 0.924 | 0.939 | 0.968 | 4.888 | 5.223 | 210.268 | 524.569 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.844 | 0.854 | 0.788 | 0.8 | 0.747 | 0.758 |
| best | 0.999 | 0.998 | 0.998 | 0.996 | 0.994 | 0.992 | |
| Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.998 | 0.997 | 0.996 | 0.961 | 0.958 |
| best | 0.999 | 0.998 | 0.997 | 0.996 | 0.961 | 0.958 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.689 | 0.71 | 0.579 | 0.604 | 0.533 | 0.557 |
| best | 0.998 | 0.997 | 0.998 | 0.996 | 0.994 | 0.992 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.919 | 0.571 | 0.879 | 0.485 |
| batch 2 | 0.843 | 0.778 | 0.773 | 0.641 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Tryptophan cluster factors {3.5} (TFClass) |
| TF family | IRF {3.5.3} (TFClass) |
| TF subfamily | {3.5.3.0} (TFClass) |
| TFClass ID | TFClass: 3.5.3.0.9 |
| HGNC | HGNC:6131 |
| MGI | MGI:107587 |
| EntrezGene (human) | GeneID:10379 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:16391 (SSTAR profile) |
| UniProt ID (human) | IRF9_HUMAN |
| UniProt ID (mouse) | IRF9_MOUSE |
| UniProt AC (human) | Q00978 (TFClass) |
| UniProt AC (mouse) | Q61179 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 3 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| PCM | IRF9.H12INVIVO.0.PSM.A.pcm |
| PWM | IRF9.H12INVIVO.0.PSM.A.pwm |
| PFM | IRF9.H12INVIVO.0.PSM.A.pfm |
| Alignment | IRF9.H12INVIVO.0.PSM.A.words.tsv |
| Threshold to P-value map | IRF9.H12INVIVO.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | IRF9.H12INVIVO.0.PSM.A_jaspar_format.txt |
| MEME format | IRF9.H12INVIVO.0.PSM.A_meme_format.meme |
| Transfac format | IRF9.H12INVIVO.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 307.0 | 83.0 | 504.0 | 100.0 |
| 02 | 175.0 | 3.0 | 789.0 | 27.0 |
| 03 | 778.0 | 8.0 | 206.0 | 2.0 |
| 04 | 988.0 | 1.0 | 4.0 | 1.0 |
| 05 | 934.0 | 16.0 | 20.0 | 24.0 |
| 06 | 336.0 | 304.0 | 208.0 | 146.0 |
| 07 | 252.0 | 184.0 | 88.0 | 470.0 |
| 08 | 52.0 | 2.0 | 935.0 | 5.0 |
| 09 | 930.0 | 3.0 | 50.0 | 11.0 |
| 10 | 983.0 | 0.0 | 7.0 | 4.0 |
| 11 | 988.0 | 0.0 | 2.0 | 4.0 |
| 12 | 33.0 | 608.0 | 348.0 | 5.0 |
| 13 | 77.0 | 270.0 | 16.0 | 631.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.309 | 0.084 | 0.507 | 0.101 |
| 02 | 0.176 | 0.003 | 0.794 | 0.027 |
| 03 | 0.783 | 0.008 | 0.207 | 0.002 |
| 04 | 0.994 | 0.001 | 0.004 | 0.001 |
| 05 | 0.94 | 0.016 | 0.02 | 0.024 |
| 06 | 0.338 | 0.306 | 0.209 | 0.147 |
| 07 | 0.254 | 0.185 | 0.089 | 0.473 |
| 08 | 0.052 | 0.002 | 0.941 | 0.005 |
| 09 | 0.936 | 0.003 | 0.05 | 0.011 |
| 10 | 0.989 | 0.0 | 0.007 | 0.004 |
| 11 | 0.994 | 0.0 | 0.002 | 0.004 |
| 12 | 0.033 | 0.612 | 0.35 | 0.005 |
| 13 | 0.077 | 0.272 | 0.016 | 0.635 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.21 | -1.083 | 0.704 | -0.9 |
| 02 | -0.348 | -3.969 | 1.151 | -2.165 |
| 03 | 1.137 | -3.248 | -0.186 | -4.207 |
| 04 | 1.375 | -4.52 | -3.777 | -4.52 |
| 05 | 1.319 | -2.647 | -2.444 | -2.275 |
| 06 | 0.3 | 0.2 | -0.177 | -0.527 |
| 07 | 0.014 | -0.298 | -1.026 | 0.634 |
| 08 | -1.538 | -4.207 | 1.32 | -3.616 |
| 09 | 1.315 | -3.969 | -1.576 | -2.979 |
| 10 | 1.37 | -4.977 | -3.356 | -3.777 |
| 11 | 1.375 | -4.977 | -4.207 | -3.777 |
| 12 | -1.975 | 0.891 | 0.335 | -3.616 |
| 13 | -1.156 | 0.082 | -2.647 | 0.928 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.44521 |
| 0.0005 | 3.75791 |
| 0.0001 | 6.47107 |