| Motif | IRF8.H12INVITRO.2.SM.B |
| Gene (human) | IRF8 (GeneCards) |
| Gene synonyms (human) | ICSBP1 |
| Gene (mouse) | Irf8 |
| Gene synonyms (mouse) | Icsbp, Icsbp1 |
| LOGO | 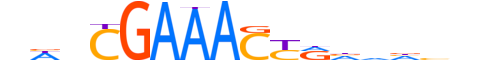 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif | IRF8.H12INVITRO.2.SM.B |
| Gene (human) | IRF8 (GeneCards) |
| Gene synonyms (human) | ICSBP1 |
| Gene (mouse) | Irf8 |
| Gene synonyms (mouse) | Icsbp, Icsbp1 |
| LOGO | 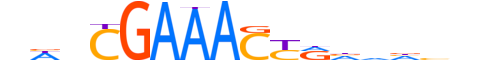 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 16 |
| Consensus | nhnCGAAACYRvhhvn |
| GC content | 45.25% |
| Information content (bits; total / per base) | 12.516 / 0.782 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9976 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 27 (157) | 0.811 | 0.906 | 0.695 | 0.821 | 0.774 | 0.857 | 2.23 | 3.053 | 150.347 | 549.585 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 7 experiments | median | 0.992 | 0.989 | 0.955 | 0.939 | 0.857 | 0.848 |
| best | 1.0 | 1.0 | 0.999 | 0.999 | 0.994 | 0.992 | |
| Methyl HT-SELEX, 2 experiments | median | 0.952 | 0.937 | 0.889 | 0.878 | 0.809 | 0.81 |
| best | 0.999 | 0.998 | 0.993 | 0.99 | 0.957 | 0.951 | |
| Non-Methyl HT-SELEX, 5 experiments | median | 0.992 | 0.989 | 0.955 | 0.939 | 0.857 | 0.848 |
| best | 1.0 | 1.0 | 0.999 | 0.999 | 0.994 | 0.992 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.925 | 0.708 | 0.878 | 0.593 |
| batch 2 | 0.845 | 0.67 | 0.808 | 0.61 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Tryptophan cluster factors {3.5} (TFClass) |
| TF family | IRF {3.5.3} (TFClass) |
| TF subfamily | {3.5.3.0} (TFClass) |
| TFClass ID | TFClass: 3.5.3.0.8 |
| HGNC | HGNC:5358 |
| MGI | MGI:96395 |
| EntrezGene (human) | GeneID:3394 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:15900 (SSTAR profile) |
| UniProt ID (human) | IRF8_HUMAN |
| UniProt ID (mouse) | IRF8_MOUSE |
| UniProt AC (human) | Q02556 (TFClass) |
| UniProt AC (mouse) | P23611 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 27 mouse |
| HT-SELEX | 5 |
| Methyl-HT-SELEX | 2 |
| PCM | IRF8.H12INVITRO.2.SM.B.pcm |
| PWM | IRF8.H12INVITRO.2.SM.B.pwm |
| PFM | IRF8.H12INVITRO.2.SM.B.pfm |
| Alignment | IRF8.H12INVITRO.2.SM.B.words.tsv |
| Threshold to P-value map | IRF8.H12INVITRO.2.SM.B.thr |
| Motif in other formats | |
| JASPAR format | IRF8.H12INVITRO.2.SM.B_jaspar_format.txt |
| MEME format | IRF8.H12INVITRO.2.SM.B_meme_format.meme |
| Transfac format | IRF8.H12INVITRO.2.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 3355.25 | 1903.25 | 1924.25 | 2793.25 |
| 02 | 5872.75 | 1221.75 | 972.75 | 1908.75 |
| 03 | 1998.0 | 2942.0 | 2060.0 | 2976.0 |
| 04 | 125.0 | 8404.0 | 218.0 | 1229.0 |
| 05 | 15.0 | 9.0 | 9950.0 | 2.0 |
| 06 | 9931.0 | 27.0 | 6.0 | 12.0 |
| 07 | 9601.0 | 0.0 | 1.0 | 374.0 |
| 08 | 9957.0 | 9.0 | 5.0 | 5.0 |
| 09 | 44.0 | 7801.0 | 2060.0 | 71.0 |
| 10 | 312.0 | 4771.0 | 226.0 | 4667.0 |
| 11 | 2701.0 | 613.0 | 5862.0 | 800.0 |
| 12 | 5118.0 | 1994.0 | 1435.0 | 1429.0 |
| 13 | 4519.0 | 1859.0 | 1632.0 | 1966.0 |
| 14 | 5204.0 | 1753.0 | 1407.0 | 1612.0 |
| 15 | 1751.25 | 4402.25 | 2167.25 | 1655.25 |
| 16 | 2040.75 | 2813.75 | 1781.75 | 3339.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.336 | 0.191 | 0.193 | 0.28 |
| 02 | 0.589 | 0.122 | 0.098 | 0.191 |
| 03 | 0.2 | 0.295 | 0.206 | 0.298 |
| 04 | 0.013 | 0.842 | 0.022 | 0.123 |
| 05 | 0.002 | 0.001 | 0.997 | 0.0 |
| 06 | 0.995 | 0.003 | 0.001 | 0.001 |
| 07 | 0.962 | 0.0 | 0.0 | 0.037 |
| 08 | 0.998 | 0.001 | 0.001 | 0.001 |
| 09 | 0.004 | 0.782 | 0.206 | 0.007 |
| 10 | 0.031 | 0.478 | 0.023 | 0.468 |
| 11 | 0.271 | 0.061 | 0.588 | 0.08 |
| 12 | 0.513 | 0.2 | 0.144 | 0.143 |
| 13 | 0.453 | 0.186 | 0.164 | 0.197 |
| 14 | 0.522 | 0.176 | 0.141 | 0.162 |
| 15 | 0.176 | 0.441 | 0.217 | 0.166 |
| 16 | 0.205 | 0.282 | 0.179 | 0.335 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.296 | -0.27 | -0.259 | 0.113 |
| 02 | 0.856 | -0.713 | -0.94 | -0.267 |
| 03 | -0.222 | 0.165 | -0.191 | 0.177 |
| 04 | -2.976 | 1.214 | -2.428 | -0.707 |
| 05 | -4.972 | -5.398 | 1.383 | -6.363 |
| 06 | 1.381 | -4.445 | -5.706 | -5.162 |
| 07 | 1.347 | -6.989 | -6.628 | -1.892 |
| 08 | 1.384 | -5.398 | -5.834 | -5.834 |
| 09 | -3.987 | 1.14 | -0.191 | -3.528 |
| 10 | -2.072 | 0.648 | -2.392 | 0.626 |
| 11 | 0.08 | -1.4 | 0.854 | -1.135 |
| 12 | 0.718 | -0.224 | -0.552 | -0.556 |
| 13 | 0.594 | -0.294 | -0.424 | -0.238 |
| 14 | 0.735 | -0.352 | -0.572 | -0.436 |
| 15 | -0.353 | 0.568 | -0.14 | -0.409 |
| 16 | -0.2 | 0.121 | -0.336 | 0.292 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.53871 |
| 0.0005 | 4.89096 |
| 0.0001 | 7.37271 |