| Motif | HTF4.H12CORE.0.PSM.A |
| Gene (human) | TCF12 (GeneCards) |
| Gene synonyms (human) | BHLHB20, HEB, HTF4 |
| Gene (mouse) | Tcf12 |
| Gene synonyms (mouse) | Alf1, Meb |
| LOGO |  |
| LOGO (reverse complement) | 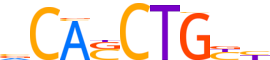 |
| Motif subtype | 0 |
| Quality | A |
| Motif | HTF4.H12CORE.0.PSM.A |
| Gene (human) | TCF12 (GeneCards) |
| Gene synonyms (human) | BHLHB20, HEB, HTF4 |
| Gene (mouse) | Tcf12 |
| Gene synonyms (mouse) | Alf1, Meb |
| LOGO |  |
| LOGO (reverse complement) | 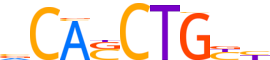 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 9 |
| Consensus | dRCAGSTGb |
| GC content | 64.2% |
| Information content (bits; total / per base) | 10.362 / 1.151 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 8 (55) | 0.843 | 0.888 | 0.705 | 0.763 | 0.797 | 0.882 | 2.187 | 2.812 | 114.824 | 286.699 |
| Mouse | 4 (28) | 0.796 | 0.841 | 0.668 | 0.731 | 0.726 | 0.788 | 2.002 | 2.281 | 107.547 | 167.284 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.959 | 0.926 | 0.955 | 0.92 | 0.9 | 0.87 |
| best | 0.96 | 0.928 | 0.958 | 0.924 | 0.925 | 0.893 | |
| Methyl HT-SELEX, 2 experiments | median | 0.959 | 0.927 | 0.957 | 0.923 | 0.915 | 0.884 |
| best | 0.96 | 0.928 | 0.958 | 0.924 | 0.925 | 0.893 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.946 | 0.907 | 0.929 | 0.889 | 0.855 | 0.825 |
| best | 0.959 | 0.926 | 0.954 | 0.919 | 0.894 | 0.866 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 3.082 | 2.293 | 0.113 | 0.064 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | E2A {1.2.1} (TFClass) |
| TF subfamily | E2A-like {1.2.1.0} (TFClass) |
| TFClass ID | TFClass: 1.2.1.0.3 |
| HGNC | HGNC:11623 |
| MGI | MGI:101877 |
| EntrezGene (human) | GeneID:6938 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21406 (SSTAR profile) |
| UniProt ID (human) | HTF4_HUMAN |
| UniProt ID (mouse) | HTF4_MOUSE |
| UniProt AC (human) | Q99081 (TFClass) |
| UniProt AC (mouse) | Q61286 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 8 human, 4 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | HTF4.H12CORE.0.PSM.A.pcm |
| PWM | HTF4.H12CORE.0.PSM.A.pwm |
| PFM | HTF4.H12CORE.0.PSM.A.pfm |
| Alignment | HTF4.H12CORE.0.PSM.A.words.tsv |
| Threshold to P-value map | HTF4.H12CORE.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | HTF4.H12CORE.0.PSM.A_jaspar_format.txt |
| MEME format | HTF4.H12CORE.0.PSM.A_meme_format.meme |
| Transfac format | HTF4.H12CORE.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 465.0 | 78.0 | 377.0 | 80.0 |
| 02 | 208.0 | 197.0 | 574.0 | 21.0 |
| 03 | 15.0 | 949.0 | 29.0 | 7.0 |
| 04 | 984.0 | 3.0 | 7.0 | 6.0 |
| 05 | 5.0 | 2.0 | 990.0 | 3.0 |
| 06 | 92.0 | 342.0 | 562.0 | 4.0 |
| 07 | 21.0 | 35.0 | 36.0 | 908.0 |
| 08 | 3.0 | 5.0 | 977.0 | 15.0 |
| 09 | 50.0 | 330.0 | 285.0 | 335.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.465 | 0.078 | 0.377 | 0.08 |
| 02 | 0.208 | 0.197 | 0.574 | 0.021 |
| 03 | 0.015 | 0.949 | 0.029 | 0.007 |
| 04 | 0.984 | 0.003 | 0.007 | 0.006 |
| 05 | 0.005 | 0.002 | 0.99 | 0.003 |
| 06 | 0.092 | 0.342 | 0.562 | 0.004 |
| 07 | 0.021 | 0.035 | 0.036 | 0.908 |
| 08 | 0.003 | 0.005 | 0.977 | 0.015 |
| 09 | 0.05 | 0.33 | 0.285 | 0.335 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.617 | -1.15 | 0.408 | -1.125 |
| 02 | -0.183 | -0.236 | 0.827 | -2.405 |
| 03 | -2.711 | 1.329 | -2.103 | -3.362 |
| 04 | 1.365 | -3.975 | -3.362 | -3.484 |
| 05 | -3.622 | -4.213 | 1.371 | -3.975 |
| 06 | -0.988 | 0.312 | 0.806 | -3.783 |
| 07 | -2.405 | -1.925 | -1.898 | 1.285 |
| 08 | -3.975 | -3.622 | 1.358 | -2.711 |
| 09 | -1.582 | 0.276 | 0.13 | 0.291 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.47315 |
| 0.0005 | 5.584545 |
| 0.0001 | 7.686795 |