| Motif | HES1.H12INVIVO.0.S.D |
| Gene (human) | HES1 (GeneCards) |
| Gene synonyms (human) | BHLHB39, HL, HRY |
| Gene (mouse) | Hes1 |
| Gene synonyms (mouse) | Hes-1 |
| LOGO | 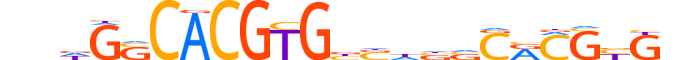 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | HES1.H12INVIVO.0.S.D |
| Gene (human) | HES1 (GeneCards) |
| Gene synonyms (human) | BHLHB39, HL, HRY |
| Gene (mouse) | Hes1 |
| Gene synonyms (mouse) | Hes-1 |
| LOGO | 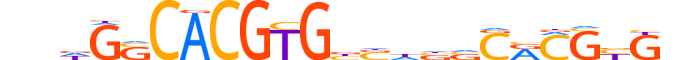 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 23 |
| Consensus | nndGSCACGTGbSdbvCRCGbGn |
| GC content | 66.76% |
| Information content (bits; total / per base) | 20.159 / 0.876 |
| Data sources | HT-SELEX |
| Aligned words | 1296 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.999 | 0.997 | 0.996 | 0.985 | 0.981 |
| best | 0.999 | 0.999 | 0.997 | 0.996 | 0.985 | 0.981 | |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | Hairy-related {1.2.4} (TFClass) |
| TF subfamily | HAIRY {1.2.4.1} (TFClass) |
| TFClass ID | TFClass: 1.2.4.1.1 |
| HGNC | HGNC:5192 |
| MGI | MGI:104853 |
| EntrezGene (human) | GeneID:3280 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:15205 (SSTAR profile) |
| UniProt ID (human) | HES1_HUMAN |
| UniProt ID (mouse) | HES1_MOUSE |
| UniProt AC (human) | Q14469 (TFClass) |
| UniProt AC (mouse) | P35428 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 0 |
| PCM | HES1.H12INVIVO.0.S.D.pcm |
| PWM | HES1.H12INVIVO.0.S.D.pwm |
| PFM | HES1.H12INVIVO.0.S.D.pfm |
| Alignment | HES1.H12INVIVO.0.S.D.words.tsv |
| Threshold to P-value map | HES1.H12INVIVO.0.S.D.thr |
| Motif in other formats | |
| JASPAR format | HES1.H12INVIVO.0.S.D_jaspar_format.txt |
| MEME format | HES1.H12INVIVO.0.S.D_meme_format.meme |
| Transfac format | HES1.H12INVIVO.0.S.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 319.0 | 345.0 | 318.0 | 314.0 |
| 02 | 456.5 | 181.5 | 346.5 | 311.5 |
| 03 | 406.0 | 73.0 | 226.0 | 591.0 |
| 04 | 61.0 | 33.0 | 1148.0 | 54.0 |
| 05 | 168.0 | 254.0 | 846.0 | 28.0 |
| 06 | 0.0 | 1296.0 | 0.0 | 0.0 |
| 07 | 1221.0 | 0.0 | 74.0 | 1.0 |
| 08 | 0.0 | 1295.0 | 1.0 | 0.0 |
| 09 | 0.0 | 0.0 | 1296.0 | 0.0 |
| 10 | 0.0 | 263.0 | 0.0 | 1033.0 |
| 11 | 0.0 | 0.0 | 1287.0 | 9.0 |
| 12 | 48.0 | 592.0 | 311.0 | 345.0 |
| 13 | 130.0 | 796.0 | 228.0 | 142.0 |
| 14 | 561.0 | 128.0 | 161.0 | 446.0 |
| 15 | 150.0 | 257.0 | 708.0 | 181.0 |
| 16 | 219.0 | 232.0 | 738.0 | 107.0 |
| 17 | 73.0 | 1076.0 | 113.0 | 34.0 |
| 18 | 782.0 | 115.0 | 358.0 | 41.0 |
| 19 | 126.0 | 1052.0 | 64.0 | 54.0 |
| 20 | 68.0 | 116.0 | 1087.0 | 25.0 |
| 21 | 60.0 | 496.0 | 139.0 | 601.0 |
| 22 | 39.75 | 93.75 | 1047.75 | 114.75 |
| 23 | 184.0 | 350.0 | 357.0 | 405.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.246 | 0.266 | 0.245 | 0.242 |
| 02 | 0.352 | 0.14 | 0.267 | 0.24 |
| 03 | 0.313 | 0.056 | 0.174 | 0.456 |
| 04 | 0.047 | 0.025 | 0.886 | 0.042 |
| 05 | 0.13 | 0.196 | 0.653 | 0.022 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 0.942 | 0.0 | 0.057 | 0.001 |
| 08 | 0.0 | 0.999 | 0.001 | 0.0 |
| 09 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 0.0 | 0.203 | 0.0 | 0.797 |
| 11 | 0.0 | 0.0 | 0.993 | 0.007 |
| 12 | 0.037 | 0.457 | 0.24 | 0.266 |
| 13 | 0.1 | 0.614 | 0.176 | 0.11 |
| 14 | 0.433 | 0.099 | 0.124 | 0.344 |
| 15 | 0.116 | 0.198 | 0.546 | 0.14 |
| 16 | 0.169 | 0.179 | 0.569 | 0.083 |
| 17 | 0.056 | 0.83 | 0.087 | 0.026 |
| 18 | 0.603 | 0.089 | 0.276 | 0.032 |
| 19 | 0.097 | 0.812 | 0.049 | 0.042 |
| 20 | 0.052 | 0.09 | 0.839 | 0.019 |
| 21 | 0.046 | 0.383 | 0.107 | 0.464 |
| 22 | 0.031 | 0.072 | 0.808 | 0.089 |
| 23 | 0.142 | 0.27 | 0.275 | 0.313 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.015 | 0.062 | -0.019 | -0.031 |
| 02 | 0.341 | -0.575 | 0.067 | -0.039 |
| 03 | 0.224 | -1.472 | -0.358 | 0.599 |
| 04 | -1.646 | -2.237 | 1.261 | -1.765 |
| 05 | -0.652 | -0.242 | 0.956 | -2.392 |
| 06 | -5.203 | 1.382 | -5.203 | -5.203 |
| 07 | 1.323 | -5.203 | -1.458 | -4.76 |
| 08 | -5.203 | 1.381 | -4.76 | -5.203 |
| 09 | -5.203 | -5.203 | 1.382 | -5.203 |
| 10 | -5.203 | -0.207 | -5.203 | 1.156 |
| 11 | -5.203 | -5.203 | 1.375 | -3.407 |
| 12 | -1.878 | 0.6 | -0.041 | 0.062 |
| 13 | -0.905 | 0.896 | -0.349 | -0.818 |
| 14 | 0.547 | -0.92 | -0.694 | 0.318 |
| 15 | -0.764 | -0.23 | 0.779 | -0.578 |
| 16 | -0.389 | -0.332 | 0.82 | -1.097 |
| 17 | -1.472 | 1.196 | -1.043 | -2.209 |
| 18 | 0.878 | -1.026 | 0.099 | -2.03 |
| 19 | -0.936 | 1.174 | -1.6 | -1.765 |
| 20 | -1.541 | -1.017 | 1.207 | -2.498 |
| 21 | -1.662 | 0.424 | -0.839 | 0.615 |
| 22 | -2.06 | -1.227 | 1.17 | -1.028 |
| 23 | -0.562 | 0.077 | 0.096 | 0.222 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.43029 |
| 0.0005 | 1.16556 |
| 0.0001 | 4.50121 |