| Motif | GLIS1.H12RSNP.0.P.B |
| Gene (human) | GLIS1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Glis1 |
| Gene synonyms (mouse) | Gli5 |
| LOGO |  |
| LOGO (reverse complement) | 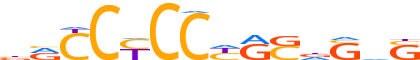 |
| Motif subtype | 0 |
| Quality | B |
| Motif | GLIS1.H12RSNP.0.P.B |
| Gene (human) | GLIS1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Glis1 |
| Gene synonyms (mouse) | Gli5 |
| LOGO |  |
| LOGO (reverse complement) | 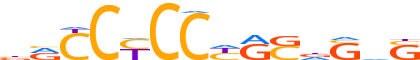 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | SdShSYRGGRGGYv |
| GC content | 72.38% |
| Information content (bits; total / per base) | 11.943 / 0.853 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (12) | 0.865 | 0.918 | 0.741 | 0.838 | 0.792 | 0.832 | 2.372 | 2.585 | 129.799 | 273.959 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.953 | 0.937 | 0.871 | 0.861 | 0.748 | 0.761 |
| best | 0.991 | 0.986 | 0.972 | 0.962 | 0.87 | 0.868 | |
| Methyl HT-SELEX, 1 experiments | median | 0.991 | 0.986 | 0.972 | 0.962 | 0.87 | 0.868 |
| best | 0.991 | 0.986 | 0.972 | 0.962 | 0.87 | 0.868 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.914 | 0.887 | 0.77 | 0.761 | 0.627 | 0.654 |
| best | 0.914 | 0.887 | 0.77 | 0.761 | 0.627 | 0.654 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.708 | 0.4 | 0.592 | 0.491 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | GLI-lile {2.3.3.1} (TFClass) |
| TFClass ID | TFClass: 2.3.3.1.4 |
| HGNC | HGNC:29525 |
| MGI | MGI:2386723 |
| EntrezGene (human) | GeneID:148979 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:230587 (SSTAR profile) |
| UniProt ID (human) | GLIS1_HUMAN |
| UniProt ID (mouse) | GLIS1_MOUSE |
| UniProt AC (human) | Q8NBF1 (TFClass) |
| UniProt AC (mouse) | Q8K1M4 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GLIS1.H12RSNP.0.P.B.pcm |
| PWM | GLIS1.H12RSNP.0.P.B.pwm |
| PFM | GLIS1.H12RSNP.0.P.B.pfm |
| Alignment | GLIS1.H12RSNP.0.P.B.words.tsv |
| Threshold to P-value map | GLIS1.H12RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | GLIS1.H12RSNP.0.P.B_jaspar_format.txt |
| MEME format | GLIS1.H12RSNP.0.P.B_meme_format.meme |
| Transfac format | GLIS1.H12RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 110.0 | 688.0 | 187.0 | 15.0 |
| 02 | 212.0 | 137.0 | 237.0 | 414.0 |
| 03 | 40.0 | 737.0 | 114.0 | 109.0 |
| 04 | 172.0 | 344.0 | 60.0 | 424.0 |
| 05 | 8.0 | 415.0 | 553.0 | 24.0 |
| 06 | 15.0 | 626.0 | 13.0 | 346.0 |
| 07 | 340.0 | 4.0 | 470.0 | 186.0 |
| 08 | 10.0 | 3.0 | 947.0 | 40.0 |
| 09 | 1.0 | 3.0 | 991.0 | 5.0 |
| 10 | 477.0 | 4.0 | 470.0 | 49.0 |
| 11 | 13.0 | 3.0 | 983.0 | 1.0 |
| 12 | 123.0 | 15.0 | 814.0 | 48.0 |
| 13 | 11.0 | 478.0 | 164.0 | 347.0 |
| 14 | 200.0 | 468.0 | 205.0 | 127.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.11 | 0.688 | 0.187 | 0.015 |
| 02 | 0.212 | 0.137 | 0.237 | 0.414 |
| 03 | 0.04 | 0.737 | 0.114 | 0.109 |
| 04 | 0.172 | 0.344 | 0.06 | 0.424 |
| 05 | 0.008 | 0.415 | 0.553 | 0.024 |
| 06 | 0.015 | 0.626 | 0.013 | 0.346 |
| 07 | 0.34 | 0.004 | 0.47 | 0.186 |
| 08 | 0.01 | 0.003 | 0.947 | 0.04 |
| 09 | 0.001 | 0.003 | 0.991 | 0.005 |
| 10 | 0.477 | 0.004 | 0.47 | 0.049 |
| 11 | 0.013 | 0.003 | 0.983 | 0.001 |
| 12 | 0.123 | 0.015 | 0.814 | 0.048 |
| 13 | 0.011 | 0.478 | 0.164 | 0.347 |
| 14 | 0.2 | 0.468 | 0.205 | 0.127 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.812 | 1.008 | -0.288 | -2.711 |
| 02 | -0.164 | -0.596 | -0.053 | 0.502 |
| 03 | -1.797 | 1.077 | -0.777 | -0.821 |
| 04 | -0.371 | 0.317 | -1.406 | 0.525 |
| 05 | -3.253 | 0.504 | 0.79 | -2.281 |
| 06 | -2.711 | 0.914 | -2.839 | 0.323 |
| 07 | 0.306 | -3.783 | 0.628 | -0.293 |
| 08 | -3.066 | -3.975 | 1.327 | -1.797 |
| 09 | -4.525 | -3.975 | 1.372 | -3.622 |
| 10 | 0.643 | -3.783 | 0.628 | -1.602 |
| 11 | -2.839 | -3.975 | 1.364 | -4.525 |
| 12 | -0.702 | -2.711 | 1.176 | -1.622 |
| 13 | -2.985 | 0.645 | -0.418 | 0.326 |
| 14 | -0.221 | 0.624 | -0.197 | -0.671 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.13331 |
| 0.0005 | 5.18251 |
| 0.0001 | 7.29971 |