| Motif | GATA2.H12RSNP.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 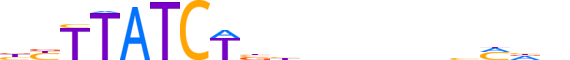 |
| Motif subtype | 1 |
| Quality | B |
| Motif | GATA2.H12RSNP.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 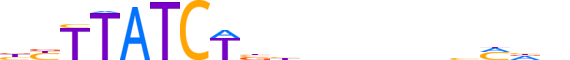 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 19 |
| Consensus | vnKbnnnnndvAGATAARv |
| GC content | 45.15% |
| Information content (bits; total / per base) | 11.704 / 0.616 |
| Data sources | ChIP-Seq |
| Aligned words | 1005 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 41 (254) | 0.835 | 0.952 | 0.7 | 0.898 | 0.823 | 0.945 | 2.567 | 4.269 | 162.915 | 316.469 |
| Mouse | 23 (136) | 0.786 | 0.955 | 0.66 | 0.898 | 0.778 | 0.954 | 2.408 | 4.266 | 127.111 | 329.237 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.907 | 0.852 | 0.877 | 0.821 | 0.824 | 0.774 |
| best | 0.919 | 0.867 | 0.896 | 0.842 | 0.853 | 0.801 | |
| Methyl HT-SELEX, 1 experiments | median | 0.919 | 0.867 | 0.896 | 0.842 | 0.853 | 0.801 |
| best | 0.919 | 0.867 | 0.896 | 0.842 | 0.853 | 0.801 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.894 | 0.837 | 0.857 | 0.799 | 0.794 | 0.747 |
| best | 0.894 | 0.837 | 0.857 | 0.799 | 0.794 | 0.747 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 4.624 | 5.538 | 0.238 | 0.129 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.636 | 0.29 | 0.615 | 0.441 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.2 |
| HGNC | HGNC:4171 |
| MGI | MGI:95662 |
| EntrezGene (human) | GeneID:2624 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14461 (SSTAR profile) |
| UniProt ID (human) | GATA2_HUMAN |
| UniProt ID (mouse) | GATA2_MOUSE |
| UniProt AC (human) | P23769 (TFClass) |
| UniProt AC (mouse) | O09100 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 41 human, 23 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GATA2.H12RSNP.1.P.B.pcm |
| PWM | GATA2.H12RSNP.1.P.B.pwm |
| PFM | GATA2.H12RSNP.1.P.B.pfm |
| Alignment | GATA2.H12RSNP.1.P.B.words.tsv |
| Threshold to P-value map | GATA2.H12RSNP.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA2.H12RSNP.1.P.B_jaspar_format.txt |
| MEME format | GATA2.H12RSNP.1.P.B_meme_format.meme |
| Transfac format | GATA2.H12RSNP.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 188.0 | 394.0 | 255.0 | 168.0 |
| 02 | 149.0 | 189.0 | 149.0 | 518.0 |
| 03 | 89.0 | 60.0 | 565.0 | 291.0 |
| 04 | 134.0 | 210.0 | 457.0 | 204.0 |
| 05 | 225.0 | 205.0 | 306.0 | 269.0 |
| 06 | 286.0 | 207.0 | 292.0 | 220.0 |
| 07 | 277.0 | 174.0 | 324.0 | 230.0 |
| 08 | 296.0 | 213.0 | 313.0 | 183.0 |
| 09 | 217.0 | 267.0 | 279.0 | 242.0 |
| 10 | 373.0 | 136.0 | 345.0 | 151.0 |
| 11 | 163.0 | 411.0 | 353.0 | 78.0 |
| 12 | 724.0 | 13.0 | 11.0 | 257.0 |
| 13 | 1.0 | 0.0 | 1004.0 | 0.0 |
| 14 | 998.0 | 4.0 | 2.0 | 1.0 |
| 15 | 4.0 | 8.0 | 10.0 | 983.0 |
| 16 | 950.0 | 3.0 | 8.0 | 44.0 |
| 17 | 856.0 | 12.0 | 114.0 | 23.0 |
| 18 | 177.0 | 160.0 | 596.0 | 72.0 |
| 19 | 370.0 | 187.0 | 385.0 | 63.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.187 | 0.392 | 0.254 | 0.167 |
| 02 | 0.148 | 0.188 | 0.148 | 0.515 |
| 03 | 0.089 | 0.06 | 0.562 | 0.29 |
| 04 | 0.133 | 0.209 | 0.455 | 0.203 |
| 05 | 0.224 | 0.204 | 0.304 | 0.268 |
| 06 | 0.285 | 0.206 | 0.291 | 0.219 |
| 07 | 0.276 | 0.173 | 0.322 | 0.229 |
| 08 | 0.295 | 0.212 | 0.311 | 0.182 |
| 09 | 0.216 | 0.266 | 0.278 | 0.241 |
| 10 | 0.371 | 0.135 | 0.343 | 0.15 |
| 11 | 0.162 | 0.409 | 0.351 | 0.078 |
| 12 | 0.72 | 0.013 | 0.011 | 0.256 |
| 13 | 0.001 | 0.0 | 0.999 | 0.0 |
| 14 | 0.993 | 0.004 | 0.002 | 0.001 |
| 15 | 0.004 | 0.008 | 0.01 | 0.978 |
| 16 | 0.945 | 0.003 | 0.008 | 0.044 |
| 17 | 0.852 | 0.012 | 0.113 | 0.023 |
| 18 | 0.176 | 0.159 | 0.593 | 0.072 |
| 19 | 0.368 | 0.186 | 0.383 | 0.063 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.288 | 0.447 | 0.015 | -0.399 |
| 02 | -0.518 | -0.282 | -0.518 | 0.72 |
| 03 | -1.025 | -1.411 | 0.807 | 0.146 |
| 04 | -0.623 | -0.178 | 0.595 | -0.207 |
| 05 | -0.11 | -0.202 | 0.196 | 0.068 |
| 06 | 0.129 | -0.192 | 0.149 | -0.132 |
| 07 | 0.097 | -0.364 | 0.253 | -0.088 |
| 08 | 0.163 | -0.164 | 0.218 | -0.314 |
| 09 | -0.145 | 0.06 | 0.104 | -0.037 |
| 10 | 0.393 | -0.608 | 0.315 | -0.505 |
| 11 | -0.429 | 0.489 | 0.338 | -1.155 |
| 12 | 1.054 | -2.844 | -2.989 | 0.022 |
| 13 | -4.53 | -4.986 | 1.38 | -4.986 |
| 14 | 1.374 | -3.788 | -4.217 | -4.53 |
| 15 | -3.788 | -3.258 | -3.071 | 1.359 |
| 16 | 1.325 | -3.98 | -3.258 | -1.711 |
| 17 | 1.221 | -2.914 | -0.782 | -2.325 |
| 18 | -0.347 | -0.447 | 0.86 | -1.233 |
| 19 | 0.385 | -0.293 | 0.424 | -1.363 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.17936 |
| 0.0005 | 5.26851 |
| 0.0001 | 7.41851 |