| Motif | GATA2.H12INVIVO.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO | 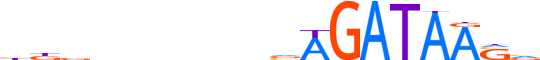 |
| LOGO (reverse complement) | 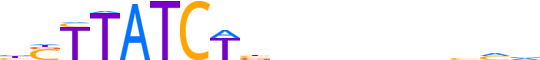 |
| Motif subtype | 1 |
| Quality | B |
| Motif | GATA2.H12INVIVO.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO | 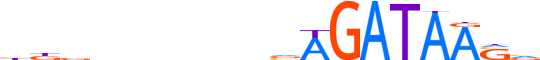 |
| LOGO (reverse complement) | 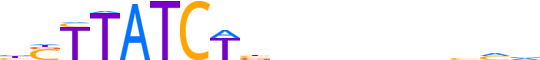 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | bdbnnnnndvAGATAASv |
| GC content | 44.76% |
| Information content (bits; total / per base) | 11.507 / 0.639 |
| Data sources | ChIP-Seq |
| Aligned words | 977 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 41 (254) | 0.843 | 0.954 | 0.716 | 0.903 | 0.828 | 0.945 | 2.613 | 4.151 | 178.616 | 332.143 |
| Mouse | 23 (136) | 0.802 | 0.959 | 0.678 | 0.904 | 0.784 | 0.955 | 2.473 | 4.178 | 140.376 | 337.658 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.913 | 0.862 | 0.884 | 0.831 | 0.836 | 0.786 |
| best | 0.924 | 0.874 | 0.901 | 0.849 | 0.862 | 0.811 | |
| Methyl HT-SELEX, 1 experiments | median | 0.924 | 0.874 | 0.901 | 0.849 | 0.862 | 0.811 |
| best | 0.924 | 0.874 | 0.901 | 0.849 | 0.862 | 0.811 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.903 | 0.85 | 0.867 | 0.812 | 0.811 | 0.762 |
| best | 0.903 | 0.85 | 0.867 | 0.812 | 0.811 | 0.762 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 3.942 | 4.58 | 0.235 | 0.113 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.65 | 0.286 | 0.616 | 0.425 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.2 |
| HGNC | HGNC:4171 |
| MGI | MGI:95662 |
| EntrezGene (human) | GeneID:2624 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14461 (SSTAR profile) |
| UniProt ID (human) | GATA2_HUMAN |
| UniProt ID (mouse) | GATA2_MOUSE |
| UniProt AC (human) | P23769 (TFClass) |
| UniProt AC (mouse) | O09100 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 41 human, 23 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GATA2.H12INVIVO.1.P.B.pcm |
| PWM | GATA2.H12INVIVO.1.P.B.pwm |
| PFM | GATA2.H12INVIVO.1.P.B.pfm |
| Alignment | GATA2.H12INVIVO.1.P.B.words.tsv |
| Threshold to P-value map | GATA2.H12INVIVO.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA2.H12INVIVO.1.P.B_jaspar_format.txt |
| MEME format | GATA2.H12INVIVO.1.P.B_meme_format.meme |
| Transfac format | GATA2.H12INVIVO.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 134.0 | 238.0 | 168.0 | 437.0 |
| 02 | 106.0 | 95.0 | 452.0 | 324.0 |
| 03 | 136.0 | 215.0 | 423.0 | 203.0 |
| 04 | 225.0 | 214.0 | 302.0 | 236.0 |
| 05 | 296.0 | 187.0 | 301.0 | 193.0 |
| 06 | 205.0 | 183.0 | 366.0 | 223.0 |
| 07 | 214.0 | 213.0 | 351.0 | 199.0 |
| 08 | 186.0 | 241.0 | 258.0 | 292.0 |
| 09 | 305.0 | 133.0 | 347.0 | 192.0 |
| 10 | 172.0 | 379.0 | 341.0 | 85.0 |
| 11 | 701.0 | 10.0 | 9.0 | 257.0 |
| 12 | 1.0 | 1.0 | 975.0 | 0.0 |
| 13 | 974.0 | 1.0 | 0.0 | 2.0 |
| 14 | 2.0 | 3.0 | 8.0 | 964.0 |
| 15 | 925.0 | 6.0 | 13.0 | 33.0 |
| 16 | 845.0 | 9.0 | 92.0 | 31.0 |
| 17 | 133.0 | 136.0 | 639.0 | 69.0 |
| 18 | 345.0 | 210.0 | 353.0 | 69.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.137 | 0.244 | 0.172 | 0.447 |
| 02 | 0.108 | 0.097 | 0.463 | 0.332 |
| 03 | 0.139 | 0.22 | 0.433 | 0.208 |
| 04 | 0.23 | 0.219 | 0.309 | 0.242 |
| 05 | 0.303 | 0.191 | 0.308 | 0.198 |
| 06 | 0.21 | 0.187 | 0.375 | 0.228 |
| 07 | 0.219 | 0.218 | 0.359 | 0.204 |
| 08 | 0.19 | 0.247 | 0.264 | 0.299 |
| 09 | 0.312 | 0.136 | 0.355 | 0.197 |
| 10 | 0.176 | 0.388 | 0.349 | 0.087 |
| 11 | 0.718 | 0.01 | 0.009 | 0.263 |
| 12 | 0.001 | 0.001 | 0.998 | 0.0 |
| 13 | 0.997 | 0.001 | 0.0 | 0.002 |
| 14 | 0.002 | 0.003 | 0.008 | 0.987 |
| 15 | 0.947 | 0.006 | 0.013 | 0.034 |
| 16 | 0.865 | 0.009 | 0.094 | 0.032 |
| 17 | 0.136 | 0.139 | 0.654 | 0.071 |
| 18 | 0.353 | 0.215 | 0.361 | 0.071 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.595 | -0.026 | -0.371 | 0.579 |
| 02 | -0.826 | -0.933 | 0.612 | 0.281 |
| 03 | -0.58 | -0.127 | 0.546 | -0.184 |
| 04 | -0.081 | -0.131 | 0.211 | -0.034 |
| 05 | 0.191 | -0.265 | 0.208 | -0.234 |
| 06 | -0.174 | -0.286 | 0.402 | -0.09 |
| 07 | -0.131 | -0.136 | 0.36 | -0.203 |
| 08 | -0.27 | -0.013 | 0.054 | 0.177 |
| 09 | 0.221 | -0.602 | 0.349 | -0.239 |
| 10 | -0.348 | 0.437 | 0.332 | -1.043 |
| 11 | 1.05 | -3.044 | -3.133 | 0.051 |
| 12 | -4.504 | -4.504 | 1.379 | -4.962 |
| 13 | 1.378 | -4.504 | -4.962 | -4.191 |
| 14 | -4.191 | -3.953 | -3.231 | 1.368 |
| 15 | 1.326 | -3.461 | -2.816 | -1.958 |
| 16 | 1.236 | -3.133 | -0.965 | -2.017 |
| 17 | -0.602 | -0.58 | 0.957 | -1.246 |
| 18 | 0.343 | -0.15 | 0.366 | -1.246 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.22851 |
| 0.0005 | 5.33836 |
| 0.0001 | 7.48216 |