| Motif | GATA2.H12INVITRO.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO | 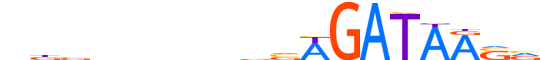 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | GATA2.H12INVITRO.1.P.B |
| Gene (human) | GATA2 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Gata2 |
| Gene synonyms (mouse) | |
| LOGO | 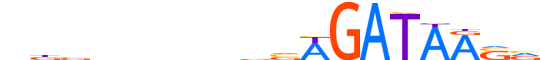 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 18 |
| Consensus | nbbnnnnndvWGATAASv |
| GC content | 47.86% |
| Information content (bits; total / per base) | 10.281 / 0.571 |
| Data sources | ChIP-Seq |
| Aligned words | 999 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 41 (254) | 0.842 | 0.953 | 0.717 | 0.907 | 0.821 | 0.938 | 2.545 | 4.111 | 168.272 | 322.26 |
| Mouse | 23 (136) | 0.808 | 0.958 | 0.697 | 0.906 | 0.797 | 0.954 | 2.6 | 4.217 | 141.745 | 330.77 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.917 | 0.867 | 0.887 | 0.834 | 0.836 | 0.788 |
| best | 0.926 | 0.878 | 0.901 | 0.851 | 0.86 | 0.81 | |
| Methyl HT-SELEX, 1 experiments | median | 0.926 | 0.878 | 0.901 | 0.851 | 0.86 | 0.81 |
| best | 0.926 | 0.878 | 0.901 | 0.851 | 0.86 | 0.81 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.908 | 0.856 | 0.872 | 0.818 | 0.813 | 0.766 |
| best | 0.908 | 0.856 | 0.872 | 0.818 | 0.813 | 0.766 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 4.741 | 5.51 | 0.233 | 0.107 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.654 | 0.281 | 0.609 | 0.423 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.2 |
| HGNC | HGNC:4171 |
| MGI | MGI:95662 |
| EntrezGene (human) | GeneID:2624 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14461 (SSTAR profile) |
| UniProt ID (human) | GATA2_HUMAN |
| UniProt ID (mouse) | GATA2_MOUSE |
| UniProt AC (human) | P23769 (TFClass) |
| UniProt AC (mouse) | O09100 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 41 human, 23 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GATA2.H12INVITRO.1.P.B.pcm |
| PWM | GATA2.H12INVITRO.1.P.B.pwm |
| PFM | GATA2.H12INVITRO.1.P.B.pfm |
| Alignment | GATA2.H12INVITRO.1.P.B.words.tsv |
| Threshold to P-value map | GATA2.H12INVITRO.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA2.H12INVITRO.1.P.B_jaspar_format.txt |
| MEME format | GATA2.H12INVITRO.1.P.B_meme_format.meme |
| Transfac format | GATA2.H12INVITRO.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 187.0 | 238.0 | 214.0 | 360.0 |
| 02 | 119.0 | 141.0 | 444.0 | 295.0 |
| 03 | 158.0 | 249.0 | 433.0 | 159.0 |
| 04 | 238.0 | 202.0 | 331.0 | 228.0 |
| 05 | 232.0 | 237.0 | 346.0 | 184.0 |
| 06 | 243.0 | 236.0 | 332.0 | 188.0 |
| 07 | 207.0 | 246.0 | 370.0 | 176.0 |
| 08 | 242.0 | 256.0 | 305.0 | 196.0 |
| 09 | 274.0 | 158.0 | 390.0 | 177.0 |
| 10 | 153.0 | 380.0 | 399.0 | 67.0 |
| 11 | 683.0 | 40.0 | 18.0 | 258.0 |
| 12 | 5.0 | 2.0 | 992.0 | 0.0 |
| 13 | 990.0 | 2.0 | 5.0 | 2.0 |
| 14 | 23.0 | 21.0 | 10.0 | 945.0 |
| 15 | 868.0 | 18.0 | 27.0 | 86.0 |
| 16 | 801.0 | 20.0 | 129.0 | 49.0 |
| 17 | 143.0 | 177.0 | 615.0 | 64.0 |
| 18 | 331.0 | 190.0 | 434.0 | 44.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.187 | 0.238 | 0.214 | 0.36 |
| 02 | 0.119 | 0.141 | 0.444 | 0.295 |
| 03 | 0.158 | 0.249 | 0.433 | 0.159 |
| 04 | 0.238 | 0.202 | 0.331 | 0.228 |
| 05 | 0.232 | 0.237 | 0.346 | 0.184 |
| 06 | 0.243 | 0.236 | 0.332 | 0.188 |
| 07 | 0.207 | 0.246 | 0.37 | 0.176 |
| 08 | 0.242 | 0.256 | 0.305 | 0.196 |
| 09 | 0.274 | 0.158 | 0.39 | 0.177 |
| 10 | 0.153 | 0.38 | 0.399 | 0.067 |
| 11 | 0.684 | 0.04 | 0.018 | 0.258 |
| 12 | 0.005 | 0.002 | 0.993 | 0.0 |
| 13 | 0.991 | 0.002 | 0.005 | 0.002 |
| 14 | 0.023 | 0.021 | 0.01 | 0.946 |
| 15 | 0.869 | 0.018 | 0.027 | 0.086 |
| 16 | 0.802 | 0.02 | 0.129 | 0.049 |
| 17 | 0.143 | 0.177 | 0.616 | 0.064 |
| 18 | 0.331 | 0.19 | 0.434 | 0.044 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.287 | -0.048 | -0.153 | 0.364 |
| 02 | -0.734 | -0.566 | 0.572 | 0.165 |
| 03 | -0.454 | -0.003 | 0.547 | -0.448 |
| 04 | -0.048 | -0.211 | 0.28 | -0.09 |
| 05 | -0.073 | -0.052 | 0.324 | -0.303 |
| 06 | -0.027 | -0.056 | 0.283 | -0.282 |
| 07 | -0.186 | -0.015 | 0.391 | -0.347 |
| 08 | -0.031 | 0.025 | 0.199 | -0.24 |
| 09 | 0.092 | -0.454 | 0.443 | -0.341 |
| 10 | -0.486 | 0.417 | 0.466 | -1.297 |
| 11 | 1.002 | -1.796 | -2.545 | 0.032 |
| 12 | -3.621 | -4.212 | 1.374 | -4.981 |
| 13 | 1.372 | -4.212 | -3.621 | -4.212 |
| 14 | -2.319 | -2.404 | -3.065 | 1.326 |
| 15 | 1.241 | -2.545 | -2.17 | -1.053 |
| 16 | 1.161 | -2.449 | -0.654 | -1.601 |
| 17 | -0.553 | -0.341 | 0.897 | -1.342 |
| 18 | 0.28 | -0.271 | 0.55 | -1.705 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.71486 |
| 0.0005 | 5.61636 |
| 0.0001 | 7.39246 |