| Motif | GATA1.H12INVIVO.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO |  |
| LOGO (reverse complement) | 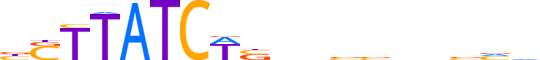 |
| Motif subtype | 0 |
| Quality | B |
| Motif | GATA1.H12INVIVO.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO |  |
| LOGO (reverse complement) | 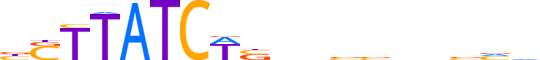 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 18 |
| Consensus | bbbnvvvvnvWGATAASv |
| GC content | 51.28% |
| Information content (bits; total / per base) | 11.989 / 0.666 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 21 (139) | 0.955 | 0.985 | 0.908 | 0.967 | 0.927 | 0.961 | 3.792 | 4.427 | 298.796 | 474.854 |
| Mouse | 37 (221) | 0.92 | 0.962 | 0.849 | 0.916 | 0.892 | 0.937 | 3.448 | 3.986 | 209.26 | 443.602 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.893 | 0.849 | 0.839 | 0.8 | 0.763 | 0.739 |
| best | 0.956 | 0.926 | 0.935 | 0.9 | 0.868 | 0.838 | |
| Methyl HT-SELEX, 1 experiments | median | 0.956 | 0.926 | 0.935 | 0.9 | 0.868 | 0.838 |
| best | 0.956 | 0.926 | 0.935 | 0.9 | 0.868 | 0.838 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.83 | 0.772 | 0.743 | 0.701 | 0.659 | 0.64 |
| best | 0.83 | 0.772 | 0.743 | 0.701 | 0.659 | 0.64 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 3.087 | 8.873 | 0.282 | 0.168 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.1 |
| HGNC | HGNC:4170 |
| MGI | MGI:95661 |
| EntrezGene (human) | GeneID:2623 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14460 (SSTAR profile) |
| UniProt ID (human) | GATA1_HUMAN |
| UniProt ID (mouse) | GATA1_MOUSE |
| UniProt AC (human) | P15976 (TFClass) |
| UniProt AC (mouse) | P17679 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 21 human, 37 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GATA1.H12INVIVO.0.P.B.pcm |
| PWM | GATA1.H12INVIVO.0.P.B.pwm |
| PFM | GATA1.H12INVIVO.0.P.B.pfm |
| Alignment | GATA1.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | GATA1.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA1.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | GATA1.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | GATA1.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 135.0 | 231.0 | 224.0 | 410.0 |
| 02 | 90.0 | 164.0 | 514.0 | 232.0 |
| 03 | 125.0 | 279.0 | 440.0 | 156.0 |
| 04 | 169.0 | 282.0 | 313.0 | 236.0 |
| 05 | 188.0 | 309.0 | 354.0 | 149.0 |
| 06 | 188.0 | 250.0 | 429.0 | 133.0 |
| 07 | 182.0 | 263.0 | 418.0 | 137.0 |
| 08 | 206.0 | 303.0 | 359.0 | 132.0 |
| 09 | 286.0 | 178.0 | 362.0 | 174.0 |
| 10 | 95.0 | 506.0 | 320.0 | 79.0 |
| 11 | 640.0 | 57.0 | 3.0 | 300.0 |
| 12 | 0.0 | 0.0 | 1000.0 | 0.0 |
| 13 | 1000.0 | 0.0 | 0.0 | 0.0 |
| 14 | 2.0 | 2.0 | 5.0 | 991.0 |
| 15 | 941.0 | 7.0 | 4.0 | 48.0 |
| 16 | 849.0 | 14.0 | 118.0 | 19.0 |
| 17 | 99.0 | 175.0 | 685.0 | 41.0 |
| 18 | 281.0 | 205.0 | 458.0 | 56.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.135 | 0.231 | 0.224 | 0.41 |
| 02 | 0.09 | 0.164 | 0.514 | 0.232 |
| 03 | 0.125 | 0.279 | 0.44 | 0.156 |
| 04 | 0.169 | 0.282 | 0.313 | 0.236 |
| 05 | 0.188 | 0.309 | 0.354 | 0.149 |
| 06 | 0.188 | 0.25 | 0.429 | 0.133 |
| 07 | 0.182 | 0.263 | 0.418 | 0.137 |
| 08 | 0.206 | 0.303 | 0.359 | 0.132 |
| 09 | 0.286 | 0.178 | 0.362 | 0.174 |
| 10 | 0.095 | 0.506 | 0.32 | 0.079 |
| 11 | 0.64 | 0.057 | 0.003 | 0.3 |
| 12 | 0.0 | 0.0 | 1.0 | 0.0 |
| 13 | 1.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.002 | 0.002 | 0.005 | 0.991 |
| 15 | 0.941 | 0.007 | 0.004 | 0.048 |
| 16 | 0.849 | 0.014 | 0.118 | 0.019 |
| 17 | 0.099 | 0.175 | 0.685 | 0.041 |
| 18 | 0.281 | 0.205 | 0.458 | 0.056 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.61 | -0.078 | -0.109 | 0.492 |
| 02 | -1.01 | -0.418 | 0.717 | -0.074 |
| 03 | -0.686 | 0.109 | 0.562 | -0.467 |
| 04 | -0.388 | 0.12 | 0.223 | -0.057 |
| 05 | -0.283 | 0.211 | 0.346 | -0.513 |
| 06 | -0.283 | 0.0 | 0.537 | -0.625 |
| 07 | -0.315 | 0.05 | 0.511 | -0.596 |
| 08 | -0.192 | 0.191 | 0.36 | -0.633 |
| 09 | 0.134 | -0.337 | 0.368 | -0.359 |
| 10 | -0.956 | 0.702 | 0.245 | -1.137 |
| 11 | 0.936 | -1.455 | -3.975 | 0.181 |
| 12 | -4.982 | -4.982 | 1.381 | -4.982 |
| 13 | 1.381 | -4.982 | -4.982 | -4.982 |
| 14 | -4.213 | -4.213 | -3.622 | 1.372 |
| 15 | 1.32 | -3.362 | -3.783 | -1.622 |
| 16 | 1.218 | -2.773 | -0.743 | -2.497 |
| 17 | -0.916 | -0.354 | 1.004 | -1.774 |
| 18 | 0.116 | -0.197 | 0.602 | -1.473 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.08866 |
| 0.0005 | 5.23246 |
| 0.0001 | 7.39551 |