| Motif | GATA1.H12INVITRO.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO | 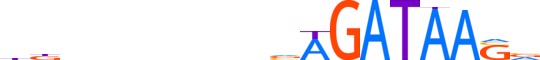 |
| LOGO (reverse complement) | 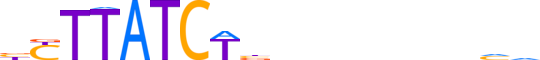 |
| Motif subtype | 0 |
| Quality | B |
| Motif | GATA1.H12INVITRO.0.P.B |
| Gene (human) | GATA1 (GeneCards) |
| Gene synonyms (human) | ERYF1, GF1 |
| Gene (mouse) | Gata1 |
| Gene synonyms (mouse) | Gf-1 |
| LOGO | 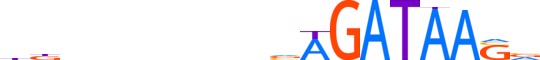 |
| LOGO (reverse complement) | 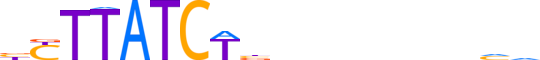 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 18 |
| Consensus | bbbnnnnnnvAGATAAvv |
| GC content | 43.08% |
| Information content (bits; total / per base) | 12.202 / 0.678 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 21 (139) | 0.941 | 0.98 | 0.88 | 0.958 | 0.927 | 0.964 | 3.743 | 4.388 | 292.041 | 446.602 |
| Mouse | 37 (221) | 0.898 | 0.96 | 0.802 | 0.915 | 0.88 | 0.941 | 3.233 | 3.894 | 191.0 | 405.699 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.903 | 0.864 | 0.848 | 0.814 | 0.775 | 0.751 |
| best | 0.964 | 0.937 | 0.946 | 0.915 | 0.876 | 0.85 | |
| Methyl HT-SELEX, 1 experiments | median | 0.964 | 0.937 | 0.946 | 0.915 | 0.876 | 0.85 |
| best | 0.964 | 0.937 | 0.946 | 0.915 | 0.876 | 0.85 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.843 | 0.792 | 0.751 | 0.712 | 0.675 | 0.652 |
| best | 0.843 | 0.792 | 0.751 | 0.712 | 0.675 | 0.652 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 2.736 | 7.087 | 0.283 | 0.183 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Other C4 zinc finger-type factors {2.2} (TFClass) |
| TF family | C4-GATA-related {2.2.1} (TFClass) |
| TF subfamily | GATA double {2.2.1.1} (TFClass) |
| TFClass ID | TFClass: 2.2.1.1.1 |
| HGNC | HGNC:4170 |
| MGI | MGI:95661 |
| EntrezGene (human) | GeneID:2623 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14460 (SSTAR profile) |
| UniProt ID (human) | GATA1_HUMAN |
| UniProt ID (mouse) | GATA1_MOUSE |
| UniProt AC (human) | P15976 (TFClass) |
| UniProt AC (mouse) | P17679 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 21 human, 37 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | GATA1.H12INVITRO.0.P.B.pcm |
| PWM | GATA1.H12INVITRO.0.P.B.pwm |
| PFM | GATA1.H12INVITRO.0.P.B.pfm |
| Alignment | GATA1.H12INVITRO.0.P.B.words.tsv |
| Threshold to P-value map | GATA1.H12INVITRO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | GATA1.H12INVITRO.0.P.B_jaspar_format.txt |
| MEME format | GATA1.H12INVITRO.0.P.B_meme_format.meme |
| Transfac format | GATA1.H12INVITRO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 150.0 | 184.0 | 221.0 | 445.0 |
| 02 | 111.0 | 130.0 | 486.0 | 273.0 |
| 03 | 147.0 | 223.0 | 385.0 | 245.0 |
| 04 | 251.0 | 184.0 | 307.0 | 258.0 |
| 05 | 268.0 | 203.0 | 297.0 | 232.0 |
| 06 | 246.0 | 184.0 | 348.0 | 222.0 |
| 07 | 250.0 | 198.0 | 322.0 | 230.0 |
| 08 | 217.0 | 236.0 | 255.0 | 292.0 |
| 09 | 312.0 | 161.0 | 303.0 | 224.0 |
| 10 | 204.0 | 404.0 | 300.0 | 92.0 |
| 11 | 711.0 | 17.0 | 5.0 | 267.0 |
| 12 | 0.0 | 0.0 | 1000.0 | 0.0 |
| 13 | 1000.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 1.0 | 999.0 |
| 15 | 964.0 | 2.0 | 1.0 | 33.0 |
| 16 | 962.0 | 3.0 | 16.0 | 19.0 |
| 17 | 139.0 | 139.0 | 694.0 | 28.0 |
| 18 | 394.0 | 188.0 | 358.0 | 60.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.15 | 0.184 | 0.221 | 0.445 |
| 02 | 0.111 | 0.13 | 0.486 | 0.273 |
| 03 | 0.147 | 0.223 | 0.385 | 0.245 |
| 04 | 0.251 | 0.184 | 0.307 | 0.258 |
| 05 | 0.268 | 0.203 | 0.297 | 0.232 |
| 06 | 0.246 | 0.184 | 0.348 | 0.222 |
| 07 | 0.25 | 0.198 | 0.322 | 0.23 |
| 08 | 0.217 | 0.236 | 0.255 | 0.292 |
| 09 | 0.312 | 0.161 | 0.303 | 0.224 |
| 10 | 0.204 | 0.404 | 0.3 | 0.092 |
| 11 | 0.711 | 0.017 | 0.005 | 0.267 |
| 12 | 0.0 | 0.0 | 1.0 | 0.0 |
| 13 | 1.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.001 | 0.999 |
| 15 | 0.964 | 0.002 | 0.001 | 0.033 |
| 16 | 0.962 | 0.003 | 0.016 | 0.019 |
| 17 | 0.139 | 0.139 | 0.694 | 0.028 |
| 18 | 0.394 | 0.188 | 0.358 | 0.06 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.506 | -0.304 | -0.122 | 0.574 |
| 02 | -0.803 | -0.648 | 0.661 | 0.087 |
| 03 | -0.526 | -0.113 | 0.429 | -0.02 |
| 04 | 0.004 | -0.304 | 0.204 | 0.031 |
| 05 | 0.069 | -0.207 | 0.171 | -0.074 |
| 06 | -0.016 | -0.304 | 0.329 | -0.118 |
| 07 | 0.0 | -0.231 | 0.252 | -0.083 |
| 08 | -0.141 | -0.057 | 0.02 | 0.154 |
| 09 | 0.22 | -0.436 | 0.191 | -0.109 |
| 10 | -0.202 | 0.477 | 0.181 | -0.988 |
| 11 | 1.041 | -2.598 | -3.622 | 0.065 |
| 12 | -4.982 | -4.982 | 1.381 | -4.982 |
| 13 | 1.381 | -4.982 | -4.982 | -4.982 |
| 14 | -4.982 | -4.982 | -4.525 | 1.38 |
| 15 | 1.345 | -4.213 | -4.525 | -1.981 |
| 16 | 1.342 | -3.975 | -2.653 | -2.497 |
| 17 | -0.582 | -0.582 | 1.017 | -2.136 |
| 18 | 0.452 | -0.283 | 0.357 | -1.406 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.67581 |
| 0.0005 | 4.96176 |
| 0.0001 | 7.55436 |