| Motif | E2F4.H12INVIVO.0.P.B |
| Gene (human) | E2F4 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | E2f4 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 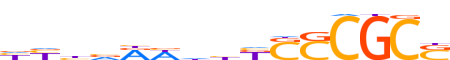 |
| Motif subtype | 0 |
| Quality | B |
| Motif | E2F4.H12INVIVO.0.P.B |
| Gene (human) | E2F4 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | E2f4 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 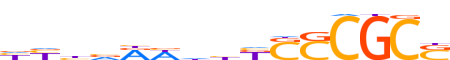 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 15 |
| Consensus | SGCGSSMddWYbdvd |
| GC content | 58.65% |
| Information content (bits; total / per base) | 9.792 / 0.653 |
| Data sources | ChIP-Seq |
| Aligned words | 999 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 8 (56) | 0.858 | 0.913 | 0.754 | 0.866 | 0.773 | 0.853 | 2.544 | 3.772 | 148.745 | 283.337 |
| Mouse | 12 (81) | 0.85 | 0.898 | 0.736 | 0.804 | 0.73 | 0.808 | 2.184 | 2.799 | 117.921 | 290.174 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.729 | 0.709 | 0.724 | 0.703 | 0.712 | 0.691 |
| best | 0.952 | 0.915 | 0.943 | 0.904 | 0.921 | 0.881 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.553 | 0.104 | 0.219 | 0.14 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Fork head/winged helix factors {3.3} (TFClass) |
| TF family | E2F {3.3.2} (TFClass) |
| TF subfamily | E2F {3.3.2.1} (TFClass) |
| TFClass ID | TFClass: 3.3.2.1.4 |
| HGNC | HGNC:3118 |
| MGI | MGI:103012 |
| EntrezGene (human) | GeneID:1874 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:104394 (SSTAR profile) |
| UniProt ID (human) | E2F4_HUMAN |
| UniProt ID (mouse) | E2F4_MOUSE |
| UniProt AC (human) | Q16254 (TFClass) |
| UniProt AC (mouse) | Q8R0K9 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 8 human, 12 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| PCM | E2F4.H12INVIVO.0.P.B.pcm |
| PWM | E2F4.H12INVIVO.0.P.B.pwm |
| PFM | E2F4.H12INVIVO.0.P.B.pfm |
| Alignment | E2F4.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | E2F4.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | E2F4.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | E2F4.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | E2F4.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 107.0 | 297.0 | 574.0 | 21.0 |
| 02 | 3.0 | 81.0 | 909.0 | 6.0 |
| 03 | 45.0 | 934.0 | 8.0 | 12.0 |
| 04 | 13.0 | 15.0 | 936.0 | 35.0 |
| 05 | 6.0 | 321.0 | 662.0 | 10.0 |
| 06 | 54.0 | 158.0 | 737.0 | 50.0 |
| 07 | 603.0 | 173.0 | 146.0 | 77.0 |
| 08 | 484.0 | 142.0 | 229.0 | 144.0 |
| 09 | 388.0 | 109.0 | 215.0 | 287.0 |
| 10 | 178.0 | 102.0 | 118.0 | 601.0 |
| 11 | 113.0 | 145.0 | 97.0 | 644.0 |
| 12 | 67.0 | 311.0 | 204.0 | 417.0 |
| 13 | 329.0 | 116.0 | 407.0 | 147.0 |
| 14 | 559.0 | 94.0 | 255.0 | 91.0 |
| 15 | 556.0 | 109.0 | 185.0 | 149.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.107 | 0.297 | 0.575 | 0.021 |
| 02 | 0.003 | 0.081 | 0.91 | 0.006 |
| 03 | 0.045 | 0.935 | 0.008 | 0.012 |
| 04 | 0.013 | 0.015 | 0.937 | 0.035 |
| 05 | 0.006 | 0.321 | 0.663 | 0.01 |
| 06 | 0.054 | 0.158 | 0.738 | 0.05 |
| 07 | 0.604 | 0.173 | 0.146 | 0.077 |
| 08 | 0.484 | 0.142 | 0.229 | 0.144 |
| 09 | 0.388 | 0.109 | 0.215 | 0.287 |
| 10 | 0.178 | 0.102 | 0.118 | 0.602 |
| 11 | 0.113 | 0.145 | 0.097 | 0.645 |
| 12 | 0.067 | 0.311 | 0.204 | 0.417 |
| 13 | 0.329 | 0.116 | 0.407 | 0.147 |
| 14 | 0.56 | 0.094 | 0.255 | 0.091 |
| 15 | 0.557 | 0.109 | 0.185 | 0.149 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.839 | 0.172 | 0.828 | -2.404 |
| 02 | -3.974 | -1.112 | 1.287 | -3.483 |
| 03 | -1.683 | 1.314 | -3.252 | -2.908 |
| 04 | -2.838 | -2.71 | 1.316 | -1.924 |
| 05 | -3.483 | 0.249 | 0.971 | -3.065 |
| 06 | -1.507 | -0.454 | 1.078 | -1.581 |
| 07 | 0.877 | -0.364 | -0.532 | -1.161 |
| 08 | 0.658 | -0.559 | -0.086 | -0.546 |
| 09 | 0.438 | -0.82 | -0.149 | 0.138 |
| 10 | -0.336 | -0.886 | -0.742 | 0.874 |
| 11 | -0.785 | -0.539 | -0.935 | 0.943 |
| 12 | -1.297 | 0.218 | -0.201 | 0.51 |
| 13 | 0.274 | -0.759 | 0.486 | -0.525 |
| 14 | 0.802 | -0.966 | 0.021 | -0.998 |
| 15 | 0.797 | -0.82 | -0.298 | -0.512 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.78536 |
| 0.0005 | 5.59071 |
| 0.0001 | 7.21601 |