| Motif | DLX1.H12RSNP.0.S.B |
| Gene (human) | DLX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Dlx1 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 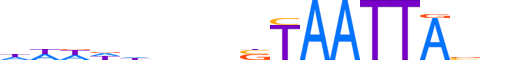 |
| Motif subtype | 0 |
| Quality | B |
| Motif | DLX1.H12RSNP.0.S.B |
| Gene (human) | DLX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Dlx1 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 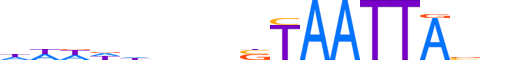 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 17 |
| Consensus | nbTAATTAvnnnhhWWh |
| GC content | 28.86% |
| Information content (bits; total / per base) | 13.214 / 0.777 |
| Data sources | HT-SELEX |
| Aligned words | 8622 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 1 (7) | 0.697 | 0.708 | 0.597 | 0.611 | 0.616 | 0.627 | 2.405 | 2.48 | 35.959 | 61.018 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 10 experiments | median | 0.974 | 0.954 | 0.967 | 0.945 | 0.95 | 0.927 |
| best | 0.99 | 0.983 | 0.984 | 0.974 | 0.973 | 0.959 | |
| Methyl HT-SELEX, 3 experiments | median | 0.974 | 0.956 | 0.966 | 0.943 | 0.945 | 0.919 |
| best | 0.977 | 0.96 | 0.972 | 0.951 | 0.964 | 0.941 | |
| Non-Methyl HT-SELEX, 7 experiments | median | 0.974 | 0.954 | 0.969 | 0.946 | 0.956 | 0.935 |
| best | 0.99 | 0.983 | 0.984 | 0.974 | 0.973 | 0.959 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.992 | 0.492 | 0.951 | 0.474 |
| batch 2 | 0.882 | 0.717 | 0.805 | 0.567 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | DLX {3.1.2.5} (TFClass) |
| TFClass ID | TFClass: 3.1.2.5.1 |
| HGNC | HGNC:2914 |
| MGI | MGI:94901 |
| EntrezGene (human) | GeneID:1745 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:13390 (SSTAR profile) |
| UniProt ID (human) | DLX1_HUMAN |
| UniProt ID (mouse) | DLX1_MOUSE |
| UniProt AC (human) | P56177 (TFClass) |
| UniProt AC (mouse) | Q64317 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 1 mouse |
| HT-SELEX | 7 |
| Methyl-HT-SELEX | 3 |
| PCM | DLX1.H12RSNP.0.S.B.pcm |
| PWM | DLX1.H12RSNP.0.S.B.pwm |
| PFM | DLX1.H12RSNP.0.S.B.pfm |
| Alignment | DLX1.H12RSNP.0.S.B.words.tsv |
| Threshold to P-value map | DLX1.H12RSNP.0.S.B.thr |
| Motif in other formats | |
| JASPAR format | DLX1.H12RSNP.0.S.B_jaspar_format.txt |
| MEME format | DLX1.H12RSNP.0.S.B_meme_format.meme |
| Transfac format | DLX1.H12RSNP.0.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1935.75 | 1629.75 | 3046.75 | 2009.75 |
| 02 | 1139.75 | 1993.75 | 3970.75 | 1517.75 |
| 03 | 0.0 | 794.0 | 0.0 | 7828.0 |
| 04 | 8622.0 | 0.0 | 0.0 | 0.0 |
| 05 | 8622.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 8622.0 |
| 07 | 0.0 | 0.0 | 0.0 | 8622.0 |
| 08 | 7256.0 | 0.0 | 1366.0 | 0.0 |
| 09 | 755.0 | 4191.0 | 3089.0 | 587.0 |
| 10 | 1191.0 | 2854.0 | 2000.0 | 2577.0 |
| 11 | 1316.0 | 2784.0 | 2276.0 | 2246.0 |
| 12 | 2737.0 | 2183.0 | 1532.0 | 2170.0 |
| 13 | 3537.0 | 1636.0 | 980.0 | 2469.0 |
| 14 | 3843.0 | 1083.0 | 682.0 | 3014.0 |
| 15 | 3637.0 | 787.0 | 450.0 | 3748.0 |
| 16 | 3426.25 | 572.25 | 571.25 | 4052.25 |
| 17 | 3216.0 | 968.0 | 858.0 | 3580.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.225 | 0.189 | 0.353 | 0.233 |
| 02 | 0.132 | 0.231 | 0.461 | 0.176 |
| 03 | 0.0 | 0.092 | 0.0 | 0.908 |
| 04 | 1.0 | 0.0 | 0.0 | 0.0 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 0.0 | 0.0 | 0.0 | 1.0 |
| 08 | 0.842 | 0.0 | 0.158 | 0.0 |
| 09 | 0.088 | 0.486 | 0.358 | 0.068 |
| 10 | 0.138 | 0.331 | 0.232 | 0.299 |
| 11 | 0.153 | 0.323 | 0.264 | 0.26 |
| 12 | 0.317 | 0.253 | 0.178 | 0.252 |
| 13 | 0.41 | 0.19 | 0.114 | 0.286 |
| 14 | 0.446 | 0.126 | 0.079 | 0.35 |
| 15 | 0.422 | 0.091 | 0.052 | 0.435 |
| 16 | 0.397 | 0.066 | 0.066 | 0.47 |
| 17 | 0.373 | 0.112 | 0.1 | 0.415 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.107 | -0.279 | 0.346 | -0.07 |
| 02 | -0.636 | -0.078 | 0.61 | -0.35 |
| 03 | -6.859 | -0.997 | -6.859 | 1.289 |
| 04 | 1.386 | -6.859 | -6.859 | -6.859 |
| 05 | 1.386 | -6.859 | -6.859 | -6.859 |
| 06 | -6.859 | -6.859 | -6.859 | 1.386 |
| 07 | -6.859 | -6.859 | -6.859 | 1.386 |
| 08 | 1.213 | -6.859 | -0.456 | -6.859 |
| 09 | -1.047 | 0.664 | 0.36 | -1.298 |
| 10 | -0.592 | 0.28 | -0.075 | 0.178 |
| 11 | -0.493 | 0.256 | 0.054 | 0.041 |
| 12 | 0.239 | 0.013 | -0.341 | 0.007 |
| 13 | 0.495 | -0.275 | -0.787 | 0.136 |
| 14 | 0.578 | -0.687 | -1.148 | 0.335 |
| 15 | 0.523 | -1.006 | -1.563 | 0.553 |
| 16 | 0.463 | -1.323 | -1.325 | 0.631 |
| 17 | 0.4 | -0.799 | -0.92 | 0.507 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.52581 |
| 0.0005 | 4.10706 |
| 0.0001 | 7.37336 |