| Motif | CUX1.H12INVIVO.2.P.C |
| Gene (human) | CUX1 (GeneCards) |
| Gene synonyms (human) | CUTL1 |
| Gene (mouse) | Cux1 |
| Gene synonyms (mouse) | Cutl1, Cux, Kiaa4047 |
| LOGO | 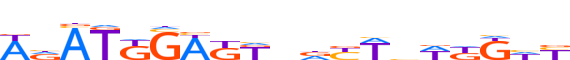 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif | CUX1.H12INVIVO.2.P.C |
| Gene (human) | CUX1 (GeneCards) |
| Gene synonyms (human) | CUTL1 |
| Gene (mouse) | Cux1 |
| Gene synonyms (mouse) | Cutl1, Cux, Kiaa4047 |
| LOGO | 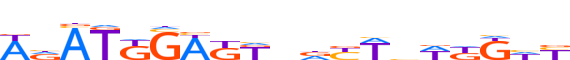 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | C |
| Motif length | 19 |
| Consensus | WdATKGWKKndYWbWKKhY |
| GC content | 41.56% |
| Information content (bits; total / per base) | 12.208 / 0.643 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (32) | 0.599 | 0.841 | 0.432 | 0.819 | 0.583 | 0.821 | 1.371 | 7.71 | 4.69 | 118.721 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.675 | 0.612 | 0.642 | 0.594 | 0.606 | 0.574 |
| best | 0.734 | 0.659 | 0.706 | 0.64 | 0.666 | 0.616 | |
| Methyl HT-SELEX, 1 experiments | median | 0.695 | 0.628 | 0.662 | 0.608 | 0.623 | 0.586 |
| best | 0.695 | 0.628 | 0.662 | 0.608 | 0.623 | 0.586 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.654 | 0.597 | 0.622 | 0.579 | 0.588 | 0.561 |
| best | 0.734 | 0.659 | 0.706 | 0.64 | 0.666 | 0.616 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.547 | 0.136 | 0.418 | 0.243 |
| batch 2 | 0.554 | 0.17 | 0.417 | 0.264 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-CUT {3.1.9} (TFClass) |
| TF subfamily | CUX {3.1.9.2} (TFClass) |
| TFClass ID | TFClass: 3.1.9.2.1 |
| HGNC | HGNC:2557 |
| MGI | MGI:88568 |
| EntrezGene (human) | GeneID:1523 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:13047 (SSTAR profile) |
| UniProt ID (human) | CUX1_HUMAN |
| UniProt ID (mouse) | CUX1_MOUSE |
| UniProt AC (human) | P39880 (TFClass) |
| UniProt AC (mouse) | P53564 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 0 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| PCM | CUX1.H12INVIVO.2.P.C.pcm |
| PWM | CUX1.H12INVIVO.2.P.C.pwm |
| PFM | CUX1.H12INVIVO.2.P.C.pfm |
| Alignment | CUX1.H12INVIVO.2.P.C.words.tsv |
| Threshold to P-value map | CUX1.H12INVIVO.2.P.C.thr |
| Motif in other formats | |
| JASPAR format | CUX1.H12INVIVO.2.P.C_jaspar_format.txt |
| MEME format | CUX1.H12INVIVO.2.P.C_meme_format.meme |
| Transfac format | CUX1.H12INVIVO.2.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 716.0 | 17.0 | 45.0 | 222.0 |
| 02 | 169.0 | 116.0 | 575.0 | 140.0 |
| 03 | 864.0 | 58.0 | 47.0 | 31.0 |
| 04 | 49.0 | 11.0 | 96.0 | 844.0 |
| 05 | 56.0 | 25.0 | 653.0 | 266.0 |
| 06 | 49.0 | 80.0 | 837.0 | 34.0 |
| 07 | 784.0 | 78.0 | 44.0 | 94.0 |
| 08 | 31.0 | 37.0 | 573.0 | 359.0 |
| 09 | 108.0 | 81.0 | 125.0 | 686.0 |
| 10 | 144.0 | 278.0 | 232.0 | 346.0 |
| 11 | 446.0 | 75.0 | 374.0 | 105.0 |
| 12 | 35.0 | 562.0 | 120.0 | 283.0 |
| 13 | 198.0 | 51.0 | 34.0 | 717.0 |
| 14 | 153.0 | 413.0 | 254.0 | 180.0 |
| 15 | 289.0 | 58.0 | 67.0 | 586.0 |
| 16 | 85.0 | 30.0 | 464.0 | 421.0 |
| 17 | 73.0 | 55.0 | 762.0 | 110.0 |
| 18 | 196.0 | 182.0 | 72.0 | 550.0 |
| 19 | 61.0 | 213.0 | 102.0 | 624.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.716 | 0.017 | 0.045 | 0.222 |
| 02 | 0.169 | 0.116 | 0.575 | 0.14 |
| 03 | 0.864 | 0.058 | 0.047 | 0.031 |
| 04 | 0.049 | 0.011 | 0.096 | 0.844 |
| 05 | 0.056 | 0.025 | 0.653 | 0.266 |
| 06 | 0.049 | 0.08 | 0.837 | 0.034 |
| 07 | 0.784 | 0.078 | 0.044 | 0.094 |
| 08 | 0.031 | 0.037 | 0.573 | 0.359 |
| 09 | 0.108 | 0.081 | 0.125 | 0.686 |
| 10 | 0.144 | 0.278 | 0.232 | 0.346 |
| 11 | 0.446 | 0.075 | 0.374 | 0.105 |
| 12 | 0.035 | 0.562 | 0.12 | 0.283 |
| 13 | 0.198 | 0.051 | 0.034 | 0.717 |
| 14 | 0.153 | 0.413 | 0.254 | 0.18 |
| 15 | 0.289 | 0.058 | 0.067 | 0.586 |
| 16 | 0.085 | 0.03 | 0.464 | 0.421 |
| 17 | 0.073 | 0.055 | 0.762 | 0.11 |
| 18 | 0.196 | 0.182 | 0.072 | 0.55 |
| 19 | 0.061 | 0.213 | 0.102 | 0.624 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1.048 | -2.598 | -1.684 | -0.118 |
| 02 | -0.388 | -0.76 | 0.829 | -0.574 |
| 03 | 1.235 | -1.439 | -1.642 | -2.04 |
| 04 | -1.602 | -2.985 | -0.946 | 1.212 |
| 05 | -1.473 | -2.243 | 0.956 | 0.062 |
| 06 | -1.602 | -1.125 | 1.204 | -1.952 |
| 07 | 1.138 | -1.15 | -1.706 | -0.967 |
| 08 | -2.04 | -1.872 | 0.826 | 0.36 |
| 09 | -0.83 | -1.113 | -0.686 | 1.005 |
| 10 | -0.547 | 0.105 | -0.074 | 0.323 |
| 11 | 0.576 | -1.188 | 0.401 | -0.858 |
| 12 | -1.925 | 0.806 | -0.727 | 0.123 |
| 13 | -0.231 | -1.563 | -1.952 | 1.049 |
| 14 | -0.487 | 0.499 | 0.016 | -0.326 |
| 15 | 0.144 | -1.439 | -1.298 | 0.848 |
| 16 | -1.066 | -2.071 | 0.615 | 0.518 |
| 17 | -1.215 | -1.49 | 1.11 | -0.812 |
| 18 | -0.241 | -0.315 | -1.228 | 0.785 |
| 19 | -1.39 | -0.159 | -0.887 | 0.911 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.22136 |
| 0.0005 | 5.10781 |
| 0.0001 | 6.98536 |