| Motif | CREB5.H12INVIVO.0.P.B |
| Gene (human) | CREB5 (GeneCards) |
| Gene synonyms (human) | CREBPA |
| Gene (mouse) | Creb5 |
| Gene synonyms (mouse) | |
| LOGO | 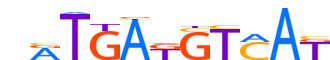 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | CREB5.H12INVIVO.0.P.B |
| Gene (human) | CREB5 (GeneCards) |
| Gene synonyms (human) | CREBPA |
| Gene (mouse) | Creb5 |
| Gene synonyms (mouse) | |
| LOGO | 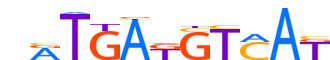 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 11 |
| Consensus | nRTGAKGTMAK |
| GC content | 36.05% |
| Information content (bits; total / per base) | 11.335 / 1.03 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (10) | 0.767 | 0.833 | 0.701 | 0.781 | 0.753 | 0.816 | 2.758 | 3.179 | 140.833 | 214.921 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.954 | 0.935 | 0.908 | 0.888 | 0.824 | 0.813 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.993 | 0.991 | |
| Methyl HT-SELEX, 2 experiments | median | 0.997 | 0.996 | 0.993 | 0.99 | 0.958 | 0.954 |
| best | 1.0 | 0.999 | 0.999 | 0.998 | 0.993 | 0.991 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.841 | 0.824 | 0.722 | 0.717 | 0.638 | 0.644 |
| best | 0.995 | 0.992 | 0.991 | 0.986 | 0.983 | 0.975 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.842 | 0.428 | 0.348 | 0.189 |
| batch 2 | 0.892 | 0.795 | 0.453 | 0.288 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | Jun-related {1.1.1} (TFClass) |
| TF subfamily | ATF2 {1.1.1.3} (TFClass) |
| TFClass ID | TFClass: 1.1.1.3.3 |
| HGNC | HGNC:16844 |
| MGI | MGI:2443973 |
| EntrezGene (human) | GeneID:9586 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:231991 (SSTAR profile) |
| UniProt ID (human) | CREB5_HUMAN |
| UniProt ID (mouse) | CREB5_MOUSE |
| UniProt AC (human) | Q02930 (TFClass) |
| UniProt AC (mouse) | Q8K1L0 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| PCM | CREB5.H12INVIVO.0.P.B.pcm |
| PWM | CREB5.H12INVIVO.0.P.B.pwm |
| PFM | CREB5.H12INVIVO.0.P.B.pfm |
| Alignment | CREB5.H12INVIVO.0.P.B.words.tsv |
| Threshold to P-value map | CREB5.H12INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | CREB5.H12INVIVO.0.P.B_jaspar_format.txt |
| MEME format | CREB5.H12INVIVO.0.P.B_meme_format.meme |
| Transfac format | CREB5.H12INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 346.0 | 159.0 | 319.0 | 176.0 |
| 02 | 662.0 | 115.0 | 181.0 | 42.0 |
| 03 | 39.0 | 6.0 | 11.0 | 944.0 |
| 04 | 3.0 | 11.0 | 806.0 | 180.0 |
| 05 | 914.0 | 29.0 | 26.0 | 31.0 |
| 06 | 76.0 | 38.0 | 263.0 | 623.0 |
| 07 | 14.0 | 117.0 | 824.0 | 45.0 |
| 08 | 21.0 | 104.0 | 27.0 | 848.0 |
| 09 | 297.0 | 667.0 | 11.0 | 25.0 |
| 10 | 970.0 | 13.0 | 2.0 | 15.0 |
| 11 | 30.0 | 117.0 | 119.0 | 734.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.346 | 0.159 | 0.319 | 0.176 |
| 02 | 0.662 | 0.115 | 0.181 | 0.042 |
| 03 | 0.039 | 0.006 | 0.011 | 0.944 |
| 04 | 0.003 | 0.011 | 0.806 | 0.18 |
| 05 | 0.914 | 0.029 | 0.026 | 0.031 |
| 06 | 0.076 | 0.038 | 0.263 | 0.623 |
| 07 | 0.014 | 0.117 | 0.824 | 0.045 |
| 08 | 0.021 | 0.104 | 0.027 | 0.848 |
| 09 | 0.297 | 0.667 | 0.011 | 0.025 |
| 10 | 0.97 | 0.013 | 0.002 | 0.015 |
| 11 | 0.03 | 0.117 | 0.119 | 0.734 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.323 | -0.449 | 0.242 | -0.348 |
| 02 | 0.97 | -0.769 | -0.32 | -1.75 |
| 03 | -1.821 | -3.484 | -2.985 | 1.324 |
| 04 | -3.975 | -2.985 | 1.166 | -0.326 |
| 05 | 1.291 | -2.103 | -2.206 | -2.04 |
| 06 | -1.175 | -1.846 | 0.05 | 0.909 |
| 07 | -2.773 | -0.752 | 1.188 | -1.684 |
| 08 | -2.405 | -0.867 | -2.171 | 1.217 |
| 09 | 0.171 | 0.977 | -2.985 | -2.243 |
| 10 | 1.351 | -2.839 | -4.213 | -2.711 |
| 11 | -2.071 | -0.752 | -0.735 | 1.073 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.375965 |
| 0.0005 | 5.31567 |
| 0.0001 | 7.21124 |