| Motif | BATF3.H12INVIVO.1.P.B |
| Gene (human) | BATF3 (GeneCards) |
| Gene synonyms (human) | SNFT |
| Gene (mouse) | Batf3 |
| Gene synonyms (mouse) | |
| LOGO | 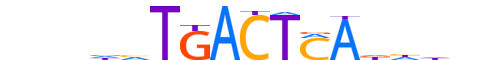 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | BATF3.H12INVIVO.1.P.B |
| Gene (human) | BATF3 (GeneCards) |
| Gene synonyms (human) | SNFT |
| Gene (mouse) | Batf3 |
| Gene synonyms (mouse) | |
| LOGO | 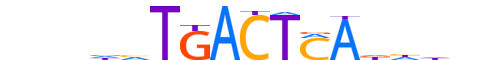 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 16 |
| Consensus | nnndvTGACTMAdhhn |
| GC content | 37.72% |
| Information content (bits; total / per base) | 11.595 / 0.725 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 4 (25) | 0.877 | 0.898 | 0.781 | 0.833 | 0.852 | 0.874 | 3.016 | 3.329 | 139.585 | 244.854 |
| Mouse | 1 (5) | 0.799 | 0.809 | 0.703 | 0.715 | 0.749 | 0.759 | 2.309 | 2.382 | 102.745 | 109.119 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.57 | 0.545 | 0.551 | 0.533 | 0.534 | 0.524 |
| best | 0.587 | 0.557 | 0.569 | 0.545 | 0.554 | 0.536 | |
| Methyl HT-SELEX, 2 experiments | median | 0.564 | 0.544 | 0.546 | 0.532 | 0.533 | 0.523 |
| best | 0.575 | 0.55 | 0.555 | 0.537 | 0.539 | 0.527 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.576 | 0.549 | 0.558 | 0.537 | 0.542 | 0.529 |
| best | 0.587 | 0.557 | 0.569 | 0.545 | 0.554 | 0.536 | |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | B-ATF-related {1.1.4} (TFClass) |
| TF subfamily | {1.1.4.0} (TFClass) |
| TFClass ID | TFClass: 1.1.4.0.3 |
| HGNC | HGNC:28915 |
| MGI | MGI:1925491 |
| EntrezGene (human) | GeneID:55509 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:381319 (SSTAR profile) |
| UniProt ID (human) | BATF3_HUMAN |
| UniProt ID (mouse) | BATF3_MOUSE |
| UniProt AC (human) | Q9NR55 (TFClass) |
| UniProt AC (mouse) | Q9D275 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 4 human, 1 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| PCM | BATF3.H12INVIVO.1.P.B.pcm |
| PWM | BATF3.H12INVIVO.1.P.B.pwm |
| PFM | BATF3.H12INVIVO.1.P.B.pfm |
| Alignment | BATF3.H12INVIVO.1.P.B.words.tsv |
| Threshold to P-value map | BATF3.H12INVIVO.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | BATF3.H12INVIVO.1.P.B_jaspar_format.txt |
| MEME format | BATF3.H12INVIVO.1.P.B_meme_format.meme |
| Transfac format | BATF3.H12INVIVO.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 296.0 | 279.0 | 181.0 | 244.0 |
| 02 | 306.0 | 168.0 | 283.0 | 243.0 |
| 03 | 366.0 | 157.0 | 191.0 | 286.0 |
| 04 | 333.0 | 78.0 | 219.0 | 370.0 |
| 05 | 500.0 | 224.0 | 144.0 | 132.0 |
| 06 | 8.0 | 5.0 | 5.0 | 982.0 |
| 07 | 10.0 | 18.0 | 815.0 | 157.0 |
| 08 | 979.0 | 6.0 | 5.0 | 10.0 |
| 09 | 1.0 | 924.0 | 0.0 | 75.0 |
| 10 | 58.0 | 5.0 | 5.0 | 932.0 |
| 11 | 124.0 | 775.0 | 85.0 | 16.0 |
| 12 | 914.0 | 14.0 | 8.0 | 64.0 |
| 13 | 218.0 | 102.0 | 223.0 | 457.0 |
| 14 | 405.0 | 234.0 | 62.0 | 299.0 |
| 15 | 281.0 | 214.0 | 106.0 | 399.0 |
| 16 | 202.0 | 344.0 | 156.0 | 298.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.296 | 0.279 | 0.181 | 0.244 |
| 02 | 0.306 | 0.168 | 0.283 | 0.243 |
| 03 | 0.366 | 0.157 | 0.191 | 0.286 |
| 04 | 0.333 | 0.078 | 0.219 | 0.37 |
| 05 | 0.5 | 0.224 | 0.144 | 0.132 |
| 06 | 0.008 | 0.005 | 0.005 | 0.982 |
| 07 | 0.01 | 0.018 | 0.815 | 0.157 |
| 08 | 0.979 | 0.006 | 0.005 | 0.01 |
| 09 | 0.001 | 0.924 | 0.0 | 0.075 |
| 10 | 0.058 | 0.005 | 0.005 | 0.932 |
| 11 | 0.124 | 0.775 | 0.085 | 0.016 |
| 12 | 0.914 | 0.014 | 0.008 | 0.064 |
| 13 | 0.218 | 0.102 | 0.223 | 0.457 |
| 14 | 0.405 | 0.234 | 0.062 | 0.299 |
| 15 | 0.281 | 0.214 | 0.106 | 0.399 |
| 16 | 0.202 | 0.344 | 0.156 | 0.298 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.168 | 0.109 | -0.32 | -0.024 |
| 02 | 0.201 | -0.394 | 0.123 | -0.028 |
| 03 | 0.379 | -0.461 | -0.267 | 0.134 |
| 04 | 0.285 | -1.15 | -0.131 | 0.39 |
| 05 | 0.69 | -0.109 | -0.547 | -0.633 |
| 06 | -3.253 | -3.622 | -3.622 | 1.363 |
| 07 | -3.066 | -2.546 | 1.177 | -0.461 |
| 08 | 1.36 | -3.484 | -3.622 | -3.066 |
| 09 | -4.525 | 1.302 | -4.982 | -1.188 |
| 10 | -1.439 | -3.622 | -3.622 | 1.311 |
| 11 | -0.694 | 1.127 | -1.066 | -2.653 |
| 12 | 1.291 | -2.773 | -3.253 | -1.343 |
| 13 | -0.136 | -0.887 | -0.113 | 0.6 |
| 14 | 0.48 | -0.066 | -1.374 | 0.178 |
| 15 | 0.116 | -0.154 | -0.849 | 0.465 |
| 16 | -0.212 | 0.317 | -0.467 | 0.175 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.24521 |
| 0.0005 | 5.25801 |
| 0.0001 | 7.34121 |