| Motif | ATF1.H12INVIVO.1.P.B |
| Gene (human) | ATF1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Atf1 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 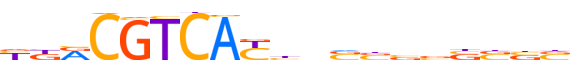 |
| Motif subtype | 1 |
| Quality | B |
| Motif | ATF1.H12INVIVO.1.P.B |
| Gene (human) | ATF1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Atf1 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 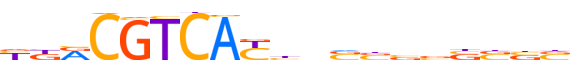 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 19 |
| Consensus | bvvvvvvbnvRTGACGYvv |
| GC content | 64.49% |
| Information content (bits; total / per base) | 12.474 / 0.657 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 5 (32) | 0.88 | 0.954 | 0.834 | 0.926 | 0.854 | 0.951 | 4.193 | 5.411 | 275.017 | 444.495 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 3.215 | 1.503 | 0.159 | 0.087 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | CREB-related {1.1.7} (TFClass) |
| TF subfamily | CREB-like {1.1.7.1} (TFClass) |
| TFClass ID | TFClass: 1.1.7.1.2 |
| HGNC | HGNC:783 |
| MGI | MGI:1298366 |
| EntrezGene (human) | GeneID:466 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:11908 (SSTAR profile) |
| UniProt ID (human) | ATF1_HUMAN |
| UniProt ID (mouse) | ATF1_MOUSE |
| UniProt AC (human) | P18846 (TFClass) |
| UniProt AC (mouse) | P81269 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 5 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| PCM | ATF1.H12INVIVO.1.P.B.pcm |
| PWM | ATF1.H12INVIVO.1.P.B.pwm |
| PFM | ATF1.H12INVIVO.1.P.B.pfm |
| Alignment | ATF1.H12INVIVO.1.P.B.words.tsv |
| Threshold to P-value map | ATF1.H12INVIVO.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | ATF1.H12INVIVO.1.P.B_jaspar_format.txt |
| MEME format | ATF1.H12INVIVO.1.P.B_meme_format.meme |
| Transfac format | ATF1.H12INVIVO.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 104.25 | 194.25 | 574.25 | 127.25 |
| 02 | 104.25 | 510.25 | 298.25 | 87.25 |
| 03 | 105.25 | 265.25 | 541.25 | 88.25 |
| 04 | 145.25 | 520.25 | 279.25 | 55.25 |
| 05 | 138.25 | 333.25 | 407.25 | 121.25 |
| 06 | 141.25 | 458.25 | 300.25 | 100.25 |
| 07 | 124.25 | 270.25 | 513.25 | 92.25 |
| 08 | 90.25 | 258.25 | 545.25 | 106.25 |
| 09 | 326.25 | 327.25 | 150.25 | 196.25 |
| 10 | 338.25 | 178.25 | 384.25 | 99.25 |
| 11 | 325.25 | 76.25 | 592.25 | 6.25 |
| 12 | 8.25 | 29.25 | 32.25 | 930.25 |
| 13 | 29.25 | 6.25 | 951.25 | 13.25 |
| 14 | 916.25 | 10.25 | 58.25 | 15.25 |
| 15 | 5.25 | 913.25 | 54.25 | 27.25 |
| 16 | 11.25 | 17.25 | 943.25 | 28.25 |
| 17 | 56.25 | 259.25 | 94.25 | 590.25 |
| 18 | 337.25 | 504.25 | 95.25 | 63.25 |
| 19 | 588.0 | 108.0 | 200.0 | 104.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.104 | 0.194 | 0.574 | 0.127 |
| 02 | 0.104 | 0.51 | 0.298 | 0.087 |
| 03 | 0.105 | 0.265 | 0.541 | 0.088 |
| 04 | 0.145 | 0.52 | 0.279 | 0.055 |
| 05 | 0.138 | 0.333 | 0.407 | 0.121 |
| 06 | 0.141 | 0.458 | 0.3 | 0.1 |
| 07 | 0.124 | 0.27 | 0.513 | 0.092 |
| 08 | 0.09 | 0.258 | 0.545 | 0.106 |
| 09 | 0.326 | 0.327 | 0.15 | 0.196 |
| 10 | 0.338 | 0.178 | 0.384 | 0.099 |
| 11 | 0.325 | 0.076 | 0.592 | 0.006 |
| 12 | 0.008 | 0.029 | 0.032 | 0.93 |
| 13 | 0.029 | 0.006 | 0.951 | 0.013 |
| 14 | 0.916 | 0.01 | 0.058 | 0.015 |
| 15 | 0.005 | 0.913 | 0.054 | 0.027 |
| 16 | 0.011 | 0.017 | 0.943 | 0.028 |
| 17 | 0.056 | 0.259 | 0.094 | 0.59 |
| 18 | 0.337 | 0.504 | 0.095 | 0.063 |
| 19 | 0.588 | 0.108 | 0.2 | 0.104 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.865 | -0.25 | 0.828 | -0.669 |
| 02 | -0.865 | 0.71 | 0.175 | -1.04 |
| 03 | -0.856 | 0.059 | 0.769 | -1.029 |
| 04 | -0.538 | 0.729 | 0.11 | -1.486 |
| 05 | -0.587 | 0.286 | 0.485 | -0.716 |
| 06 | -0.566 | 0.603 | 0.182 | -0.904 |
| 07 | -0.692 | 0.077 | 0.716 | -0.985 |
| 08 | -1.007 | 0.032 | 0.776 | -0.846 |
| 09 | 0.265 | 0.268 | -0.505 | -0.24 |
| 10 | 0.301 | -0.336 | 0.427 | -0.913 |
| 11 | 0.262 | -1.172 | 0.858 | -3.452 |
| 12 | -3.228 | -2.095 | -2.003 | 1.309 |
| 13 | -2.095 | -3.452 | 1.331 | -2.822 |
| 14 | 1.294 | -3.045 | -1.434 | -2.696 |
| 15 | -3.586 | 1.291 | -1.503 | -2.162 |
| 16 | -2.965 | -2.585 | 1.323 | -2.128 |
| 17 | -1.468 | 0.036 | -0.964 | 0.855 |
| 18 | 0.298 | 0.698 | -0.954 | -1.354 |
| 19 | 0.851 | -0.83 | -0.221 | -0.867 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.08141 |
| 0.0005 | 5.04656 |
| 0.0001 | 7.05921 |