| Motif | AP2E.H12INVITRO.0.SM.B |
| Gene (human) | TFAP2E (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tfap2e |
| Gene synonyms (mouse) | Tcfap2e |
| LOGO |  |
| LOGO (reverse complement) | 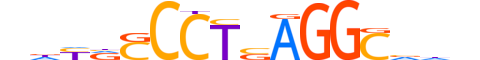 |
| Motif subtype | 0 |
| Quality | B |
| Motif | AP2E.H12INVITRO.0.SM.B |
| Gene (human) | TFAP2E (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tfap2e |
| Gene synonyms (mouse) | Tcfap2e |
| LOGO |  |
| LOGO (reverse complement) | 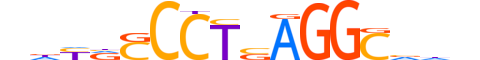 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 16 |
| Consensus | ndbGCCTSAGGSbRhn |
| GC content | 61.62% |
| Information content (bits; total / per base) | 14.474 / 0.905 |
| Data sources | HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 9965 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.998 | 0.997 | 0.997 | 0.995 | 0.994 | 0.991 |
| best | 0.999 | 0.999 | 0.999 | 0.998 | 0.997 | 0.996 | |
| Methyl HT-SELEX, 1 experiments | median | 0.999 | 0.999 | 0.999 | 0.998 | 0.997 | 0.996 |
| best | 0.999 | 0.999 | 0.999 | 0.998 | 0.997 | 0.996 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.997 | 0.995 | 0.995 | 0.993 | 0.991 | 0.986 |
| best | 0.997 | 0.995 | 0.995 | 0.993 | 0.991 | 0.986 | |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-span-helix factors (bHSH) {1.3} (TFClass) |
| TF family | AP-2 {1.3.1} (TFClass) |
| TF subfamily | {1.3.1.0} (TFClass) |
| TFClass ID | TFClass: 1.3.1.0.5 |
| HGNC | HGNC:30774 |
| MGI | MGI:2679630 |
| EntrezGene (human) | GeneID:339488 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:332937 (SSTAR profile) |
| UniProt ID (human) | AP2E_HUMAN |
| UniProt ID (mouse) | AP2E_MOUSE |
| UniProt AC (human) | Q6VUC0 (TFClass) |
| UniProt AC (mouse) | Q6VUP9 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| PCM | AP2E.H12INVITRO.0.SM.B.pcm |
| PWM | AP2E.H12INVITRO.0.SM.B.pwm |
| PFM | AP2E.H12INVITRO.0.SM.B.pfm |
| Alignment | AP2E.H12INVITRO.0.SM.B.words.tsv |
| Threshold to P-value map | AP2E.H12INVITRO.0.SM.B.thr |
| Motif in other formats | |
| JASPAR format | AP2E.H12INVITRO.0.SM.B_jaspar_format.txt |
| MEME format | AP2E.H12INVITRO.0.SM.B_meme_format.meme |
| Transfac format | AP2E.H12INVITRO.0.SM.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 3023.75 | 2071.75 | 2288.75 | 2580.75 |
| 02 | 2755.0 | 1056.0 | 1896.0 | 4258.0 |
| 03 | 1501.0 | 1915.0 | 1517.0 | 5032.0 |
| 04 | 188.0 | 1598.0 | 8035.0 | 144.0 |
| 05 | 3.0 | 9957.0 | 4.0 | 1.0 |
| 06 | 1.0 | 9867.0 | 9.0 | 88.0 |
| 07 | 9.0 | 985.0 | 6.0 | 8965.0 |
| 08 | 536.0 | 4414.0 | 4320.0 | 695.0 |
| 09 | 8526.0 | 35.0 | 1401.0 | 3.0 |
| 10 | 663.0 | 3.0 | 9296.0 | 3.0 |
| 11 | 6.0 | 3.0 | 9951.0 | 5.0 |
| 12 | 170.0 | 3261.0 | 6467.0 | 67.0 |
| 13 | 1205.0 | 5866.0 | 1292.0 | 1602.0 |
| 14 | 5722.0 | 589.0 | 2797.0 | 857.0 |
| 15 | 2505.25 | 2092.25 | 734.25 | 4633.25 |
| 16 | 3134.5 | 1520.5 | 3006.5 | 2303.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.303 | 0.208 | 0.23 | 0.259 |
| 02 | 0.276 | 0.106 | 0.19 | 0.427 |
| 03 | 0.151 | 0.192 | 0.152 | 0.505 |
| 04 | 0.019 | 0.16 | 0.806 | 0.014 |
| 05 | 0.0 | 0.999 | 0.0 | 0.0 |
| 06 | 0.0 | 0.99 | 0.001 | 0.009 |
| 07 | 0.001 | 0.099 | 0.001 | 0.9 |
| 08 | 0.054 | 0.443 | 0.434 | 0.07 |
| 09 | 0.856 | 0.004 | 0.141 | 0.0 |
| 10 | 0.067 | 0.0 | 0.933 | 0.0 |
| 11 | 0.001 | 0.0 | 0.999 | 0.001 |
| 12 | 0.017 | 0.327 | 0.649 | 0.007 |
| 13 | 0.121 | 0.589 | 0.13 | 0.161 |
| 14 | 0.574 | 0.059 | 0.281 | 0.086 |
| 15 | 0.251 | 0.21 | 0.074 | 0.465 |
| 16 | 0.315 | 0.153 | 0.302 | 0.231 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.194 | -0.184 | -0.085 | 0.035 |
| 02 | 0.101 | -0.857 | -0.273 | 0.536 |
| 03 | -0.506 | -0.263 | -0.495 | 0.703 |
| 04 | -2.573 | -0.444 | 1.17 | -2.836 |
| 05 | -6.153 | 1.385 | -5.981 | -6.627 |
| 06 | -6.627 | 1.376 | -5.397 | -3.318 |
| 07 | -5.397 | -0.926 | -5.705 | 1.28 |
| 08 | -1.533 | 0.572 | 0.55 | -1.274 |
| 09 | 1.23 | -4.202 | -0.575 | -6.153 |
| 10 | -1.321 | -6.153 | 1.316 | -6.153 |
| 11 | -5.705 | -6.153 | 1.384 | -5.833 |
| 12 | -2.672 | 0.269 | 0.953 | -3.583 |
| 13 | -0.725 | 0.856 | -0.656 | -0.441 |
| 14 | 0.831 | -1.439 | 0.116 | -1.065 |
| 15 | 0.006 | -0.174 | -1.219 | 0.62 |
| 16 | 0.229 | -0.493 | 0.188 | -0.078 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.10266 |
| 0.0005 | 3.64306 |
| 0.0001 | 6.72261 |