| Motif | ANDR.H12RSNP.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 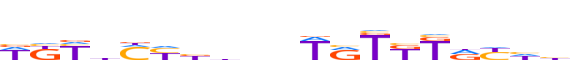 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif | ANDR.H12RSNP.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 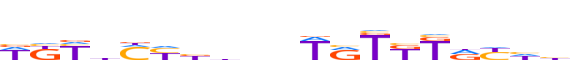 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 19 |
| Consensus | WKYhMYbbnnWRTKKdYhb |
| GC content | 41.91% |
| Information content (bits; total / per base) | 8.445 / 0.444 |
| Data sources | ChIP-Seq |
| Aligned words | 976 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 237 (413) | 0.725 | 0.851 | 0.624 | 0.814 | 0.786 | 0.892 | 3.084 | 4.984 | 64.921 | 380.222 |
| Mouse | 20 (35) | 0.557 | 0.764 | 0.416 | 0.754 | 0.663 | 0.821 | 1.986 | 4.738 | 19.357 | 107.523 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 3 experiments | median | 0.536 | 0.521 | 0.529 | 0.517 | 0.519 | 0.513 |
| best | 0.579 | 0.554 | 0.554 | 0.538 | 0.536 | 0.527 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.57 | 13.767 | 0.05 | 0.022 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Steroid hormone receptors {2.1.1} (TFClass) |
| TF subfamily | GR-like (NR3C) {2.1.1.1} (TFClass) |
| TFClass ID | TFClass: 2.1.1.1.4 |
| HGNC | HGNC:644 |
| MGI | MGI:88064 |
| EntrezGene (human) | GeneID:367 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:11835 (SSTAR profile) |
| UniProt ID (human) | ANDR_HUMAN |
| UniProt ID (mouse) | ANDR_MOUSE |
| UniProt AC (human) | P10275 (TFClass) |
| UniProt AC (mouse) | P19091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 237 human, 20 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 0 |
| PCM | ANDR.H12RSNP.2.P.B.pcm |
| PWM | ANDR.H12RSNP.2.P.B.pwm |
| PFM | ANDR.H12RSNP.2.P.B.pfm |
| Alignment | ANDR.H12RSNP.2.P.B.words.tsv |
| Threshold to P-value map | ANDR.H12RSNP.2.P.B.thr |
| Motif in other formats | |
| JASPAR format | ANDR.H12RSNP.2.P.B_jaspar_format.txt |
| MEME format | ANDR.H12RSNP.2.P.B_meme_format.meme |
| Transfac format | ANDR.H12RSNP.2.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 165.0 | 72.0 | 114.0 | 625.0 |
| 02 | 60.0 | 112.0 | 639.0 | 165.0 |
| 03 | 89.0 | 125.0 | 81.0 | 681.0 |
| 04 | 170.0 | 211.0 | 162.0 | 433.0 |
| 05 | 123.0 | 671.0 | 69.0 | 113.0 |
| 06 | 132.0 | 365.0 | 16.0 | 463.0 |
| 07 | 137.0 | 147.0 | 196.0 | 496.0 |
| 08 | 129.0 | 188.0 | 262.0 | 397.0 |
| 09 | 211.0 | 231.0 | 256.0 | 278.0 |
| 10 | 208.0 | 344.0 | 180.0 | 244.0 |
| 11 | 158.0 | 33.0 | 55.0 | 730.0 |
| 12 | 206.0 | 106.0 | 613.0 | 51.0 |
| 13 | 47.0 | 64.0 | 67.0 | 798.0 |
| 14 | 47.0 | 96.0 | 211.0 | 622.0 |
| 15 | 40.0 | 60.0 | 145.0 | 731.0 |
| 16 | 340.0 | 68.0 | 484.0 | 84.0 |
| 17 | 74.0 | 548.0 | 63.0 | 291.0 |
| 18 | 266.0 | 204.0 | 57.0 | 449.0 |
| 19 | 122.0 | 271.0 | 186.0 | 397.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.169 | 0.074 | 0.117 | 0.64 |
| 02 | 0.061 | 0.115 | 0.655 | 0.169 |
| 03 | 0.091 | 0.128 | 0.083 | 0.698 |
| 04 | 0.174 | 0.216 | 0.166 | 0.444 |
| 05 | 0.126 | 0.688 | 0.071 | 0.116 |
| 06 | 0.135 | 0.374 | 0.016 | 0.474 |
| 07 | 0.14 | 0.151 | 0.201 | 0.508 |
| 08 | 0.132 | 0.193 | 0.268 | 0.407 |
| 09 | 0.216 | 0.237 | 0.262 | 0.285 |
| 10 | 0.213 | 0.352 | 0.184 | 0.25 |
| 11 | 0.162 | 0.034 | 0.056 | 0.748 |
| 12 | 0.211 | 0.109 | 0.628 | 0.052 |
| 13 | 0.048 | 0.066 | 0.069 | 0.818 |
| 14 | 0.048 | 0.098 | 0.216 | 0.637 |
| 15 | 0.041 | 0.061 | 0.149 | 0.749 |
| 16 | 0.348 | 0.07 | 0.496 | 0.086 |
| 17 | 0.076 | 0.561 | 0.065 | 0.298 |
| 18 | 0.273 | 0.209 | 0.058 | 0.46 |
| 19 | 0.125 | 0.278 | 0.191 | 0.407 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.388 | -1.204 | -0.753 | 0.936 |
| 02 | -1.382 | -0.77 | 0.958 | -0.388 |
| 03 | -0.996 | -0.662 | -1.089 | 1.022 |
| 04 | -0.358 | -0.144 | -0.406 | 0.571 |
| 05 | -0.678 | 1.007 | -1.245 | -0.762 |
| 06 | -0.608 | 0.4 | -2.629 | 0.637 |
| 07 | -0.572 | -0.502 | -0.217 | 0.706 |
| 08 | -0.631 | -0.259 | 0.071 | 0.484 |
| 09 | -0.144 | -0.054 | 0.048 | 0.13 |
| 10 | -0.158 | 0.341 | -0.302 | 0.0 |
| 11 | -0.431 | -1.957 | -1.466 | 1.091 |
| 12 | -0.168 | -0.825 | 0.917 | -1.539 |
| 13 | -1.618 | -1.319 | -1.274 | 1.18 |
| 14 | -1.618 | -0.922 | -0.144 | 0.932 |
| 15 | -1.773 | -1.382 | -0.516 | 1.093 |
| 16 | 0.33 | -1.26 | 0.681 | -1.053 |
| 17 | -1.177 | 0.805 | -1.334 | 0.175 |
| 18 | 0.086 | -0.178 | -1.431 | 0.607 |
| 19 | -0.686 | 0.104 | -0.269 | 0.484 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.82306 |
| 0.0005 | 5.51866 |
| 0.0001 | 6.98876 |