| Motif | ANDR.H12INVITRO.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 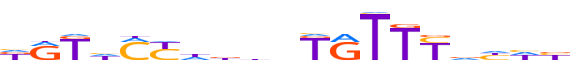 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif | ANDR.H12INVITRO.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 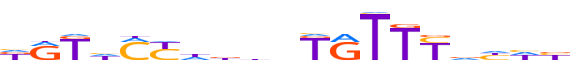 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 19 |
| Consensus | dRKhMYhbdnKRTTYvhhb |
| GC content | 37.72% |
| Information content (bits; total / per base) | 10.753 / 0.566 |
| Data sources | ChIP-Seq |
| Aligned words | 801 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 237 (413) | 0.727 | 0.864 | 0.618 | 0.817 | 0.795 | 0.915 | 2.969 | 4.927 | 73.602 | 409.721 |
| Mouse | 20 (35) | 0.589 | 0.77 | 0.434 | 0.745 | 0.649 | 0.807 | 1.834 | 4.154 | 26.456 | 113.119 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 3 experiments | median | 0.566 | 0.537 | 0.551 | 0.531 | 0.534 | 0.523 |
| best | 0.791 | 0.714 | 0.75 | 0.684 | 0.692 | 0.646 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.472 | 10.427 | 0.045 | 0.02 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Steroid hormone receptors {2.1.1} (TFClass) |
| TF subfamily | GR-like (NR3C) {2.1.1.1} (TFClass) |
| TFClass ID | TFClass: 2.1.1.1.4 |
| HGNC | HGNC:644 |
| MGI | MGI:88064 |
| EntrezGene (human) | GeneID:367 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:11835 (SSTAR profile) |
| UniProt ID (human) | ANDR_HUMAN |
| UniProt ID (mouse) | ANDR_MOUSE |
| UniProt AC (human) | P10275 (TFClass) |
| UniProt AC (mouse) | P19091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 237 human, 20 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 0 |
| PCM | ANDR.H12INVITRO.2.P.B.pcm |
| PWM | ANDR.H12INVITRO.2.P.B.pwm |
| PFM | ANDR.H12INVITRO.2.P.B.pfm |
| Alignment | ANDR.H12INVITRO.2.P.B.words.tsv |
| Threshold to P-value map | ANDR.H12INVITRO.2.P.B.thr |
| Motif in other formats | |
| JASPAR format | ANDR.H12INVITRO.2.P.B_jaspar_format.txt |
| MEME format | ANDR.H12INVITRO.2.P.B_meme_format.meme |
| Transfac format | ANDR.H12INVITRO.2.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 126.0 | 87.0 | 139.0 | 449.0 |
| 02 | 164.0 | 38.0 | 542.0 | 57.0 |
| 03 | 70.0 | 22.0 | 92.0 | 617.0 |
| 04 | 176.0 | 111.0 | 106.0 | 408.0 |
| 05 | 139.0 | 579.0 | 15.0 | 68.0 |
| 06 | 54.0 | 442.0 | 8.0 | 297.0 |
| 07 | 315.0 | 97.0 | 81.0 | 308.0 |
| 08 | 104.0 | 140.0 | 190.0 | 367.0 |
| 09 | 162.0 | 126.0 | 187.0 | 326.0 |
| 10 | 126.0 | 250.0 | 149.0 | 276.0 |
| 11 | 64.0 | 37.0 | 69.0 | 631.0 |
| 12 | 236.0 | 22.0 | 535.0 | 8.0 |
| 13 | 19.0 | 19.0 | 7.0 | 756.0 |
| 14 | 17.0 | 18.0 | 82.0 | 684.0 |
| 15 | 11.0 | 187.0 | 51.0 | 552.0 |
| 16 | 331.0 | 138.0 | 235.0 | 97.0 |
| 17 | 139.0 | 412.0 | 47.0 | 203.0 |
| 18 | 200.0 | 137.0 | 46.0 | 418.0 |
| 19 | 76.0 | 169.0 | 129.0 | 427.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.157 | 0.109 | 0.174 | 0.561 |
| 02 | 0.205 | 0.047 | 0.677 | 0.071 |
| 03 | 0.087 | 0.027 | 0.115 | 0.77 |
| 04 | 0.22 | 0.139 | 0.132 | 0.509 |
| 05 | 0.174 | 0.723 | 0.019 | 0.085 |
| 06 | 0.067 | 0.552 | 0.01 | 0.371 |
| 07 | 0.393 | 0.121 | 0.101 | 0.385 |
| 08 | 0.13 | 0.175 | 0.237 | 0.458 |
| 09 | 0.202 | 0.157 | 0.233 | 0.407 |
| 10 | 0.157 | 0.312 | 0.186 | 0.345 |
| 11 | 0.08 | 0.046 | 0.086 | 0.788 |
| 12 | 0.295 | 0.027 | 0.668 | 0.01 |
| 13 | 0.024 | 0.024 | 0.009 | 0.944 |
| 14 | 0.021 | 0.022 | 0.102 | 0.854 |
| 15 | 0.014 | 0.233 | 0.064 | 0.689 |
| 16 | 0.413 | 0.172 | 0.293 | 0.121 |
| 17 | 0.174 | 0.514 | 0.059 | 0.253 |
| 18 | 0.25 | 0.171 | 0.057 | 0.522 |
| 19 | 0.095 | 0.211 | 0.161 | 0.533 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.458 | -0.823 | -0.361 | 0.803 |
| 02 | -0.198 | -1.627 | 0.99 | -1.236 |
| 03 | -1.036 | -2.144 | -0.768 | 1.12 |
| 04 | -0.128 | -0.583 | -0.629 | 0.707 |
| 05 | -0.361 | 1.056 | -2.494 | -1.064 |
| 06 | -1.288 | 0.787 | -3.039 | 0.391 |
| 07 | 0.45 | -0.716 | -0.893 | 0.428 |
| 08 | -0.648 | -0.354 | -0.052 | 0.602 |
| 09 | -0.21 | -0.458 | -0.068 | 0.484 |
| 10 | -0.458 | 0.22 | -0.293 | 0.319 |
| 11 | -1.123 | -1.653 | -1.05 | 1.142 |
| 12 | 0.163 | -2.144 | 0.978 | -3.039 |
| 13 | -2.279 | -2.279 | -3.148 | 1.322 |
| 14 | -2.381 | -2.329 | -0.881 | 1.223 |
| 15 | -2.769 | -0.068 | -1.344 | 1.009 |
| 16 | 0.499 | -0.369 | 0.159 | -0.716 |
| 17 | -0.361 | 0.717 | -1.423 | 0.014 |
| 18 | -0.001 | -0.376 | -1.444 | 0.732 |
| 19 | -0.955 | -0.168 | -0.435 | 0.753 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.56141 |
| 0.0005 | 5.39541 |
| 0.0001 | 7.13346 |