| Motif | ANDR.H12CORE.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 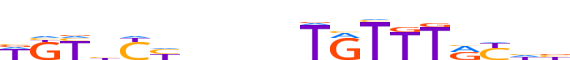 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif | ANDR.H12CORE.2.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO | 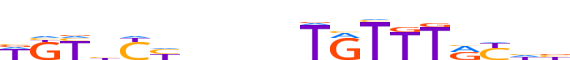 |
| LOGO (reverse complement) |  |
| Motif subtype | 2 |
| Quality | B |
| Motif length | 19 |
| Consensus | WKYhYhbnnnTRTTTRYhb |
| GC content | 40.77% |
| Information content (bits; total / per base) | 11.495 / 0.605 |
| Data sources | ChIP-Seq |
| Aligned words | 1004 |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 237 (413) | 0.757 | 0.88 | 0.665 | 0.839 | 0.803 | 0.909 | 3.255 | 5.246 | 75.678 | 413.131 |
| Mouse | 20 (35) | 0.566 | 0.78 | 0.432 | 0.774 | 0.656 | 0.83 | 2.023 | 4.797 | 14.31 | 106.699 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 3 experiments | median | 0.542 | 0.525 | 0.532 | 0.519 | 0.524 | 0.515 |
| best | 0.569 | 0.546 | 0.547 | 0.532 | 0.533 | 0.523 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.529 | 12.345 | 0.052 | 0.022 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Steroid hormone receptors {2.1.1} (TFClass) |
| TF subfamily | GR-like (NR3C) {2.1.1.1} (TFClass) |
| TFClass ID | TFClass: 2.1.1.1.4 |
| HGNC | HGNC:644 |
| MGI | MGI:88064 |
| EntrezGene (human) | GeneID:367 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:11835 (SSTAR profile) |
| UniProt ID (human) | ANDR_HUMAN |
| UniProt ID (mouse) | ANDR_MOUSE |
| UniProt AC (human) | P10275 (TFClass) |
| UniProt AC (mouse) | P19091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 237 human, 20 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 0 |
| PCM | ANDR.H12CORE.2.P.B.pcm |
| PWM | ANDR.H12CORE.2.P.B.pwm |
| PFM | ANDR.H12CORE.2.P.B.pfm |
| Alignment | ANDR.H12CORE.2.P.B.words.tsv |
| Threshold to P-value map | ANDR.H12CORE.2.P.B.thr |
| Motif in other formats | |
| JASPAR format | ANDR.H12CORE.2.P.B_jaspar_format.txt |
| MEME format | ANDR.H12CORE.2.P.B_meme_format.meme |
| Transfac format | ANDR.H12CORE.2.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 145.0 | 79.0 | 110.0 | 670.0 |
| 02 | 71.0 | 78.0 | 755.0 | 100.0 |
| 03 | 88.25 | 100.25 | 53.25 | 762.25 |
| 04 | 177.25 | 200.25 | 150.25 | 476.25 |
| 05 | 74.25 | 740.25 | 37.25 | 152.25 |
| 06 | 104.25 | 386.25 | 50.25 | 463.25 |
| 07 | 174.25 | 214.25 | 213.25 | 402.25 |
| 08 | 184.25 | 225.25 | 249.25 | 345.25 |
| 09 | 202.25 | 210.25 | 266.25 | 325.25 |
| 10 | 198.25 | 316.25 | 153.25 | 336.25 |
| 11 | 62.25 | 21.25 | 33.25 | 887.25 |
| 12 | 183.25 | 52.25 | 759.25 | 9.25 |
| 13 | 33.25 | 48.25 | 17.25 | 905.25 |
| 14 | 17.25 | 25.25 | 144.25 | 817.25 |
| 15 | 20.25 | 14.25 | 108.25 | 861.25 |
| 16 | 390.25 | 68.25 | 504.25 | 41.25 |
| 17 | 64.25 | 643.25 | 41.25 | 255.25 |
| 18 | 328.25 | 183.25 | 65.25 | 427.25 |
| 19 | 84.25 | 288.25 | 172.25 | 459.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.144 | 0.079 | 0.11 | 0.667 |
| 02 | 0.071 | 0.078 | 0.752 | 0.1 |
| 03 | 0.088 | 0.1 | 0.053 | 0.759 |
| 04 | 0.177 | 0.199 | 0.15 | 0.474 |
| 05 | 0.074 | 0.737 | 0.037 | 0.152 |
| 06 | 0.104 | 0.385 | 0.05 | 0.461 |
| 07 | 0.174 | 0.213 | 0.212 | 0.401 |
| 08 | 0.184 | 0.224 | 0.248 | 0.344 |
| 09 | 0.201 | 0.209 | 0.265 | 0.324 |
| 10 | 0.197 | 0.315 | 0.153 | 0.335 |
| 11 | 0.062 | 0.021 | 0.033 | 0.884 |
| 12 | 0.183 | 0.052 | 0.756 | 0.009 |
| 13 | 0.033 | 0.048 | 0.017 | 0.902 |
| 14 | 0.017 | 0.025 | 0.144 | 0.814 |
| 15 | 0.02 | 0.014 | 0.108 | 0.858 |
| 16 | 0.389 | 0.068 | 0.502 | 0.041 |
| 17 | 0.064 | 0.641 | 0.041 | 0.254 |
| 18 | 0.327 | 0.183 | 0.065 | 0.426 |
| 19 | 0.084 | 0.287 | 0.172 | 0.457 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.544 | -1.141 | -0.816 | 0.978 |
| 02 | -1.246 | -1.154 | 1.097 | -0.91 |
| 03 | -1.033 | -0.908 | -1.525 | 1.106 |
| 04 | -0.345 | -0.224 | -0.509 | 0.637 |
| 05 | -1.202 | 1.077 | -1.869 | -0.496 |
| 06 | -0.869 | 0.429 | -1.581 | 0.61 |
| 07 | -0.362 | -0.157 | -0.162 | 0.469 |
| 08 | -0.307 | -0.107 | -0.007 | 0.317 |
| 09 | -0.214 | -0.176 | 0.059 | 0.258 |
| 10 | -0.234 | 0.23 | -0.489 | 0.291 |
| 11 | -1.374 | -2.398 | -1.978 | 1.258 |
| 12 | -0.312 | -1.544 | 1.102 | -3.136 |
| 13 | -1.978 | -1.621 | -2.589 | 1.278 |
| 14 | -2.589 | -2.237 | -0.549 | 1.176 |
| 15 | -2.442 | -2.761 | -0.832 | 1.228 |
| 16 | 0.439 | -1.284 | 0.694 | -1.772 |
| 17 | -1.343 | 0.937 | -1.772 | 0.017 |
| 18 | 0.267 | -0.312 | -1.328 | 0.529 |
| 19 | -1.078 | 0.137 | -0.373 | 0.601 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.37516 |
| 0.0005 | 5.24491 |
| 0.0001 | 7.07456 |